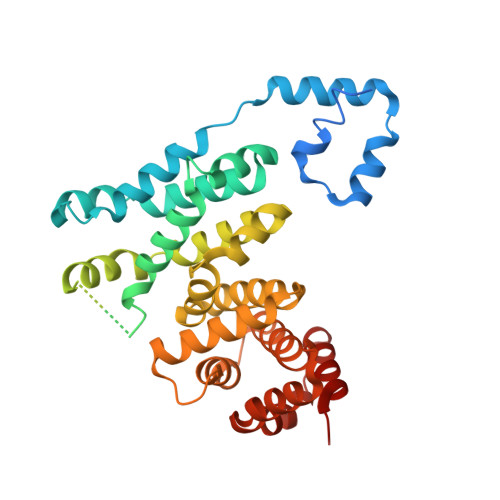
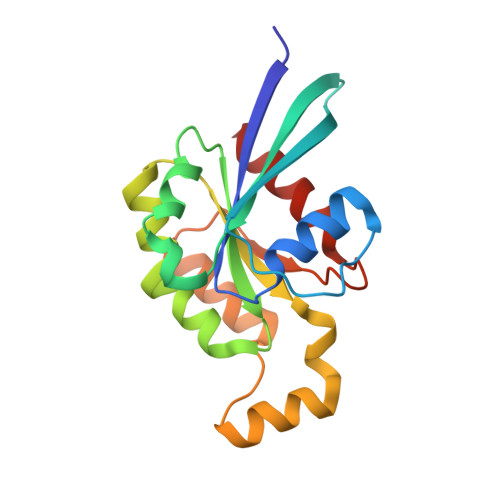
The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation.
Kuhn, S., Erdmann, C., Kage, F., Block, J., Schwenkmezger, L., Steffen, A., Rottner, K., Geyer, M.(2015) Nat Commun 6: 7088-7088
- PubMed: 25963737
- DOI: https://doi.org/10.1038/ncomms8088
- Primary Citation of Related Structures:
4YC7, 4YDH - PubMed Abstract:
Formins are actin polymerization factors that elongate unbranched actin filaments at the barbed end. Rho family GTPases activate Diaphanous-related formins through the relief of an autoregulatory interaction. The crystal structures of the N-terminal domains of human FMNL1 and FMNL2 in complex with active Cdc42 show that Cdc42 mediates contacts with all five armadillo repeats of the formin with specific interactions formed by the Rho-GTPase insert helix. Mutation of three residues within Rac1 results in a gain-of-function mutation for FMNL2 binding and reconstitution of the Cdc42 phenotype in vivo. Dimerization of FMNL1 through a parallel coiled coil segment leads to formation of an umbrella-shaped structure that—together with Cdc42—spans more than 15 nm in diameter. The two interacting FMNL-Cdc42 heterodimers expose six membrane interaction motifs on a convex protein surface, the assembly of which may facilitate actin filament elongation at the leading edge of lamellipodia and filopodia.
Organizational Affiliation:
1] Center of Advanced European Studies and Research, Group Physical Biochemistry, Ludwig-Erhard-Allee 2, Bonn 53175, Germany [2] Department of Physical Biochemistry, Max Planck Institute of Molecular Physiology, Otto-Hahn-Strasse 11, Dortmund 44227, Germany.