E. coli acetyl-CoA carboxylase, narrow helical local reconstruction, 3.18 Angstrom
Xu, X., Silva de Sousa, A., Boram, T.J., Jiang, W., Lohman, R.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha | 316 | Escherichia coli | Mutation(s): 0 Gene Names: accA, b0185, JW0180 EC: 2.1.3.15 | 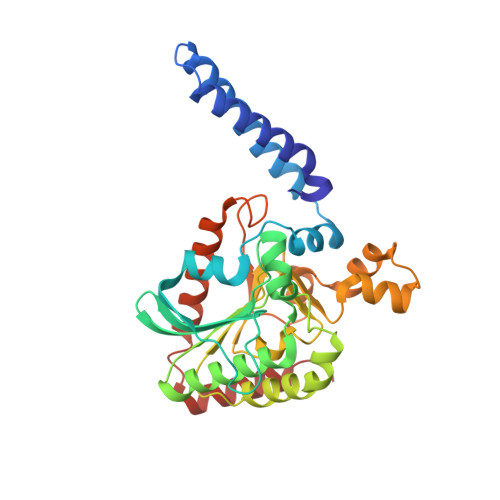 | |
UniProt | |||||
Find proteins for P0ABD5 (Escherichia coli (strain K12)) Explore P0ABD5 Go to UniProtKB: P0ABD5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ABD5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Biotin carboxyl carrier protein of acetyl-CoA carboxylase | 77 | Escherichia coli | Mutation(s): 0 Gene Names: accB, fabE, b3255, JW3223 | 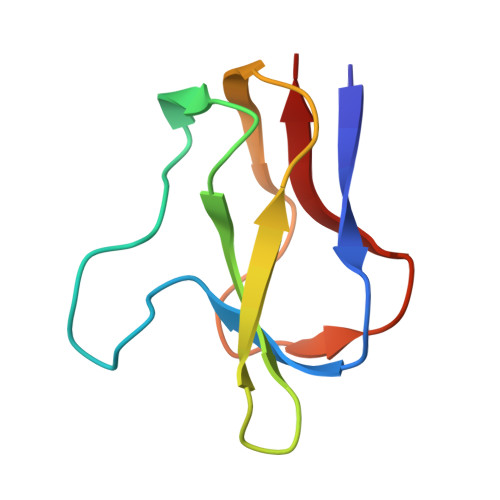 | |
UniProt | |||||
Find proteins for P0ABD8 (Escherichia coli (strain K12)) Explore P0ABD8 Go to UniProtKB: P0ABD8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ABD8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Biotin carboxylase | 446 | Escherichia coli | Mutation(s): 0 Gene Names: accC, fabG, b3256, JW3224 EC: 6.3.4.14 | 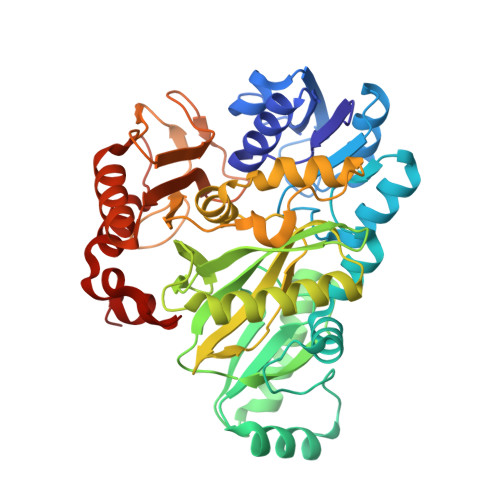 | |
UniProt | |||||
Find proteins for P24182 (Escherichia coli (strain K12)) Explore P24182 Go to UniProtKB: P24182 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P24182 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta | 284 | Escherichia coli | Mutation(s): 0 Gene Names: accD, dedB, usg, b2316, JW2313 EC: 2.1.3.15 | 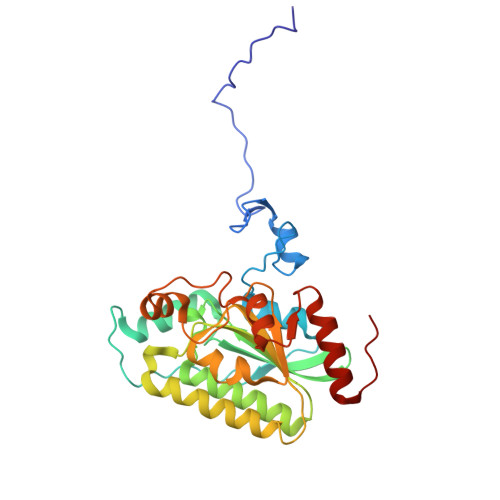 | |
UniProt | |||||
Find proteins for P0A9Q5 (Escherichia coli (strain K12)) Explore P0A9Q5 Go to UniProtKB: P0A9Q5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A9Q5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ACO Query on ACO | N [auth D], S [auth H] | ACETYL COENZYME *A C23 H38 N7 O17 P3 S ZSLZBFCDCINBPY-ZSJPKINUSA-N |  | ||
| ADP Query on ADP | K [auth C], P [auth G] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| BTN Query on BTN | J [auth B], O [auth F] | BIOTIN C10 H16 N2 O3 S YBJHBAHKTGYVGT-ZKWXMUAHSA-N |  | ||
| ZN Query on ZN | M [auth D], R [auth H] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | L [auth C], Q [auth G] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM140290 |