2002 PDB News
Contents:
Read the latest PDB news. Earlier news is available and is archived in the RCSB PDB newsletters.
23-Dec-2002
Happy Holidays from the PDB!
The PDB staff wish to extend our best wishes to the community for a happy holiday season and a wonderful new year!
17-Dec-2002
PDB Update Schedule for December 23-31
The PDB update that would normally occur on December 24 will instead take place on December 23. The update that would normally occur on December 31 will take place on December 30.
The regular PDB update schedule will resume with the January 7 update.
PDB to be Highlighted on New Jersey Network News
A segment highlighting the PDB will be shown as part of the New Jersey Network (NJN) News on Friday, December 27, 2002. The show will air at 6:00PM, 7:30PM, and 11:00PM on NJN Public Television (check your local listings for channel information) and at 5:30 PM on WNET/New York (Channel 13).
The news is also broadcast online at http://www.njn.net/television/webcast/.
10-Dec-2002
PDB CD-ROM Set #102 and Subsequent Releases
Issue 102 of the PDB CD-ROM sets is now shipping. With this release, 18,796 experimentally determined structures from the PDB FTP site as of October 1, 2002 are contained on a 5 CD-ROM set. The theoretical structures are also included in this set in a models directory.
The experimental data, X-ray structure factors and NMR constraints, are available as separate products for the structures for which they were deposited. For additional information and ordering instructions refer to the CD-ROM page at http://www.rcsb.org/pdb/cdrom.html.
The January 2003 release of the PDB CD-ROM sets, issue 103, will be a full release of experimentally determined structures. Starting with the April 2003 release an incremental set, structures released since the January 2003 release, will be sent to subscribers. Structures re-released for any reason will be included in the update.
July and October releases will be updates also. New subscribers will receive the January release and all subsequent updates. A list of files that have become obsolete since the last update will be sent with each release so users can update their set of structures.
The index files in the pub/resource sub-directory will continue to include all structures in the current PDB FTP site as of that release.
Experimental data, NMR constraints and X-ray structure factors, will be handled in the same manner as the structures - a complete set in January and incremental updates for the three subsequent quarters.
3-Dec-2002
New PDB Mirror Site in Germany
A new RCSB PDB mirror site has been established at the Max Delbr�ck Center for Molecular Medicine in Berlin, Germany. This site is now accessible at http://www.pdb.mdc-berlin.de/. A complete list of the RCSB PDB's eight worldwide mirror sites can be found at http://www.rcsb.org/pdb/mirrors.html.
26-Nov-2002
PDB Focus: Sequence Prerelease
PDB depositors are given the opportunity to prerelease a sequence in advance of the coordinates. ADIT's default setting is set to sequence prerelease.
From the PDB status search at http://www.rcsb.org/pdb/status.html, users may query all available sequences, or query based on criteria such as title or deposition date.
This feature was developed in response to requests made to the PDB. It is hoped that the prerelease of sequence data will prevent unintended duplication of effort in structure determination. It will allow users to conduct blind tests of structure prediction and modeling techniques.
Rsync Script for FTP Mirroring
An rsync script, rsyncPDB.sh, has been made available at ftp://ftp.rcsb.org/pub/pdb/software/. This script assists users in setting up their own local mirrors of the PDB FTP site. Before successfully running it, users will need to set three variables in rsyncPDB.sh to suit their local setup. The rsync script may be preferred to the mirror.pl script that was previously recommended for local mirroring of the FTP site. Questions about the rsync script may be sent to [email protected].
19-Nov-2002

PDB Annual Report 2002 Now Available
The PDB Annual Report 2002 is now being distributed. This document features a detailed look at the third full year of the operation of the PDB by Rutgers, the State University of New Jersey; the San Diego Supercomputer Center at the University of California, San Diego; and the Center for Advanced Research in Biotechnology of the National Institute of Standards and Technology -- three members of the RCSB. It highlights accomplishments during this period, from July 1, 2001 through June 30, 2002, and describes future developments of the PDB resource.
Requests for printed copies of the PDB Annual Report can be sent to [email protected]. This document is also available on-line in PDF format at http://www.rcsb.org/pdb/annual_report02.pdf.
12-Nov-2002
PDB Art Exhibit part of CCMB's Silver Jubilee Celebrations and Symposium
The PDB's "Art of Science" exhibit will be part of the Centre for Cellular and Molecular Biology's (CCMB) Silver Jubilee Celebrations and Symposium on "The Current Excitement in Biology" in Hyderabad, India.
The exhibit will be on display at the CCMB campus's main building from November 15, 2002 through February 28, 2003. The Symposium (http://www.ccmb.res.in/symposium/) will take place November 24-29, 2002.
The PDB would like to see this exhibit travel to other places. If you would be interested in sponsoring this exhibit at your institution, please let us know at [email protected].
05-Nov-2002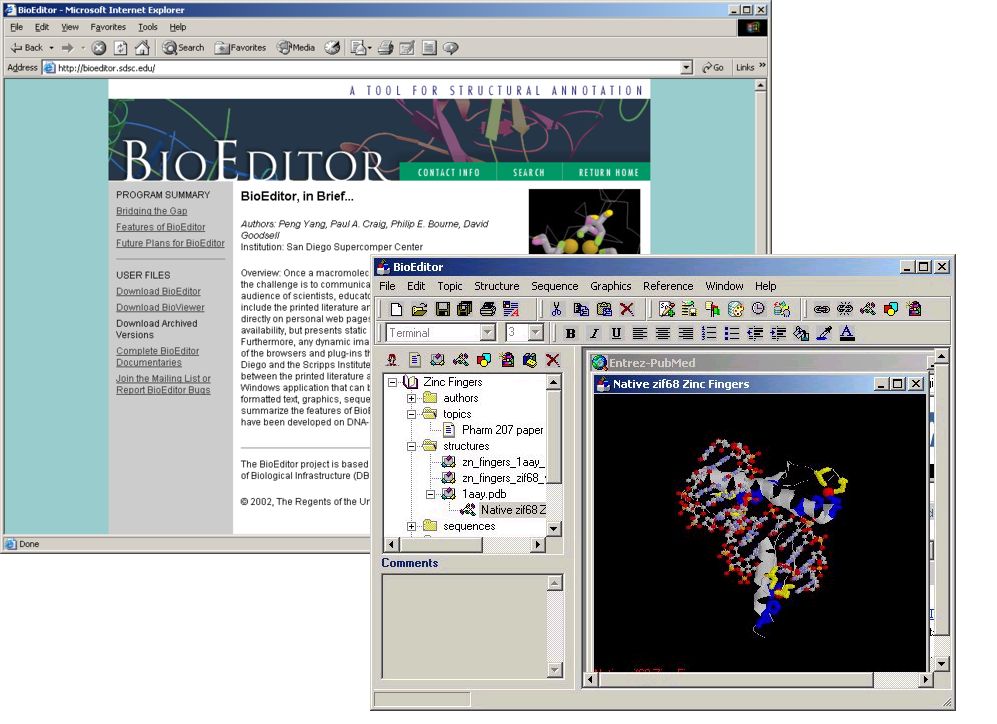
BioEditor Now Available from the PDB Web Site
BioEditor, a program for creating and viewing structure presentations, is now accessible from the PDB Web site at http://www.rcsb.org/pdb/education.html#Other and http://www.rcsb.org/pdb/software-list.html#Graphics.
BioEditor (http://bioeditor.sdsc.edu/) is a tool to bridge the gap between printed literature and current Web-based presentation formats for macromolecular structures. It is a standalone Windows application that can be used to prepare and present structure annotations containing formatted text, graphics, sequence data, and interactive molecular views--all in a single document or set of documents. BioEditor facilitates the communication of structure data to a diverse audience by allowing users to create and view dynamic content in a uniform format that can be widely distributed through the internet.
BioEditor is designed to be used by structural scientists reporting and evaluating their data, as well as by educators and students who are seeking to relate structure to function in biological macromolecules. The BioEditor application includes features that enable the user to enter data, images, and references. Many features also link directly to resources on the internet.
Complete BioEditor documentaries on the PDB structures of a zinc binuclear cluster and the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) enzyme are accessible from the Protein Documentaries section of PDB's Education Page at http://www.rcsb.org/pdb/education.html#Documentaries.
29-Oct-2002
Theoretical Models Search Interface on the PDB Beta Web Site
A simple search interface for theoretical model structures is now available from the PDB Beta Web Site at http://beta.rcsb.org/pdb/cgi/models.cgi. This interface facilitates searches by PDB ID, author, and compound, and is accessible from the top of the SearchLite and SearchFields interfaces. Feedback on this new feature may be sent to [email protected].
22-Oct-2002
"Banking on Structures": BioIT World article looks at the PDB
The latest issue of BioIT World examines all aspects of the PDB in "Banking on Structures" by Tracy Smith Schmidt at http://www.bio-itworld.com/archive/100902/banking.html.
15-Oct-2002
PDB Newsletter 15 Released
The Fall 2002 issue of the PDB Newsletter is now available in HTML format at http://www.rcsb.org/pdb/newsletter/2002q3/. This document describes PDB's activities over the past three months in the areas of data deposition and processing; structural genomics; data query, reporting, and access; and outreach. This issue will soon be available in .pdf and print format as well. Subscriptions for the quarterly printed distribution of the PDB newsletter may be requested by sending your postal address to [email protected].
08-Oct-2002
PDB Paper Published in Bioinformatics
The latest issue of Bioinformatics contains a paper describing the Ontology Driven Architecture that was developed to increase efficiency in the use of macromolecular structure (MMS) data contained in the PDB. The paper describes the OpenMMS Toolkit, a suite of software tools that conform to the OMG/LSR Corba standard (OMG specification formal/02-05-01). The OpenMMS Toolkit, is based on a metamodel architecture and is written entirely in Java. It contains an mmCIF parser, reference and database Corba servers, a relational database loader and a prototype XML formatted file converter. Detailed documentation and all of the source code are available at http://openmms.sdsc.edu.
D.S. Greer, J.D. Westbrook, P.E. Bourne (2002): An ontology driven architecture for derived representations of macromolecular structure. Bioinformatics, 18(9), pp. 1280-1281. © Oxford University Press.
01-Oct-2002
| Dihydrofolate Reductase, the Molecule of the Month for October, 2002. |
PDB Focus: Educational Resources at the PDB
The "Educational Resources" page and the "Molecule of the Month" series are two important resources that the PDB maintains to serve the user community.
Each month, Dr. David S. Goodsell (The Scripps Research Institute) highlights a key biological molecule with illustrations and text designed for a general audience. These features are available from the PDB home page, and are archived at http://www.rcsb.org/pdb/molecules/molecule_list.html.
The PDB Educational Resources page at http://www.rcsb.org/pdb/education.html compiles molecular biology resources for audiences ranging from elementary level students to undergraduates to the general public. Proteins and nucleic acid tutorials, PDB articles and animations, protein documentaries, the World Index of Molecular Visualization Resources, and an illustrated glossary of crystallographic and NMR terminology are a few of the resources linked to from this page. Suggestions for additions to this page are appreciated and can be sent to [email protected].
24-Sep-2002
PDB Focus: PDBOBS
The PDBOBS database at http://pdbobs.sdsc.edu/index.cgi archives versions of PDB entries that have been obsoleted or superseded by more recent versions. These entries are made available for historical purposes, and to allow new versions of software to be tested against the same data sets as earlier versions of the software. The PDBOBS search interface can be used to locate obsolete entries by PDB ID or keyword. A list of obsolete PDB ID's is also available for browsing.
The result of all queries and the links in the browsing pages will graphically display the history of all versions of the PDB entry as well as a textual comparison of the key features of the structure.
A simple view of the sequence and backbone of any obsolete entry can be seen through the QuickPDB applet linked from the PDBOBS home page. This feature also includes a simple search by PDB ID capability. Content statistics and a list of enhancements are also accessible from the PDBOBS home page.
Obsolete PDB entries are also available from the FTP Web site at ftp://ftp.rcsb.org/pub/pdb/data/structures/obsolete/.
17-Sep-2002
CD-ROM News: Experimental Data To Be Offered as Separate CD-ROM Set
The number of entries in the PDB archive is ever-increasing, and this is reflected in the number of CD-ROMs required to contain each quarterly release. In order to reduce the number of disks in each set, beginning with the October 2002 CD-ROM release, subscribers will receive only the coordinate files for structure entries. This will include structures solved by X-Ray and NMR experimental techniques as well as those determined theoretically.
The experimental data files will no longer be included automatically. This will reduce the number of CD-ROMs to be loaded by 50%. Requests to receive either the X-Ray structure factors or NMR constraint files can be submitted at http://www.nist.gov/srd/o_nist801.htm.
All recipients of the current release, July 2002, received a flyer with the CD set that can be returned to request the experimental data as well as update their address. That same flyer is included as a README file on the first CD-ROM disk in directory pub and can thus be attached to an email to use in requesting the experimental data.
All CD-ROM services will continue to be available free of cost.
10-Sep-2002
Experimental Data Reporting and Model Access Enhancements
Enhancements have been made to the PDB site for reporting the availability of experimental data and for accessing model data.
The availability of experimental data for a particular structure can now be included in reports generated for a set of structures resulting from a PDB search. This new column indicates whether there is a structure factor or NMR restraint file available for each structure. This feature is included in the Structure Summary report and is offered as an option for the customizable reports. Tabular reports of results can be saved in HTML format or plain text format; the latter can also be imported into a spreadsheet program such as Excel.
Links to the models directory on the PDB FTP site (ftp://ftp.rcsb.org/pub/pdb/data/structures/models/) have been added to the SearchLite and SearchFields pages to facilitate access to these entries from the PDB Web site. This directory includes links to released theoretical model files, a set of index files containing the keyword, author, source organism, compound, resolution, crystal dimensions for these files, and the obsoleted theoretical model files.
Limited search capability for theoretical models will be made available in the near future.
03-Sep-2002
PDB Focus: Author Searches
Users can query for a particular author of a structure or a primary citation using the SearchFields interface at http://www.rcsb.org/pdb/cgi/queryForm.cgi. In the Citation Author field, enter the two initials and last name of the target author. An example of an author query would be:
S.S.TaylorThe initials should be separated by "."s without spaces between them or the last name. If the query is not formed this way, no results will be returned. For authors with a title included in their name, the format should be:
W.E.Royer Jr.
H.Brumer III
A search can also be performed using the author's last name only, for example:
MatthewsThis will return all authors with the last name "Matthews" that are referenced in the PDB archive, as listed in the JRNL records or PDB REMARK 1 records in the structure entries. To remove a particular author from this result list, select the Refine Your Query option from the pull down menu at the top of the Query Result Browser page, which will return the SearchFields page. At the top, select the BUTNOT logic button and enter the author to be removed from the results list in the Citation Author field. For example:
B.W.Matthewswill remove all entries that reference "B.W.Matthews" from the "Matthews" result list.
Multiple authors can be queried for by performing a search for a single author as noted above, then selecting Refine Your Query from the pull down menu at the top of the results page, and entering the second author in the Citation Author field (note: the AND logic button on the SearchFields page must be selected, which is the default).
The Text Search field can also be used to query for part or all of one or more author names in a single query, such as:
Taylor and ZhengHowever, users should be aware that this type of search scans the entire text of each PDB file and will return entries that include text that literally matches the query, so some undesired results may occur.
Requests for assistance with queries may be sent to [email protected].
27-Aug-2002
PDB at ISMB, IUCr, and the Protein Society Symposium
The PDB thanks everyone who visited our exhibit booths at several meetings earlier this month: the Intelligent Systems for Molecular Biology (ISMB) conference in Edmonton, Canada, the Protein Society's Annual Symposium in San Diego, CA, and the XIX Congress and General Assembly of the International Union of Crystallography (IUCr) in Geneva, Switzerland.
The PDB Users Lunch held at the IUCr meeting set the stage for feedback from our users and a very productive discussion. We would like to thank our sponsors for their support at this lunch: Area Detector Systems Corporation, Blake Industries, Inc., deCODE genetics Emerald BioStructures Products, GlaxoSmithKline, Hampton Research, IBM, Merck Co., Inc. - USA, Proctor & Gamble Pharmaceuticals, and the Schering-Plough Research Institute.
20-Aug-2002
New mmCIF and Data Processing Software Available
RCSB-developed programs for mmCIF and data processing -- including MAXIT, PDB_EXTRACT, and an mmCIF database loader -- are now available from the PDB's software page for download. These programs are available in source and binary versions.
RCSB-developed applications available for use with mmCIF and for data processing include:
- CIFTr -- An application program for translating files in mmCIF format into files in PDB format
- PDB Validation Suite -- A tool for processing and checking structure data
- MAXIT -- An application for processing and curation of macromolecular structure data
- ADIT -- A package for editing and checking structure data entries
- PDB_EXTRACT -- Tools and examples for extracting mmCIF data from structure determination applications
- mmCIF Loader -- An application to load mmCIF data into relational databases and XML
The PDB's Software page is a portal to software developed by the RCSB and others in the macromolecular structure community. Links to external software resources relating to mmCIF, crystallography, NMR, structure analysis and verification, modeling and simulation, and molecular graphics are also available here.
This page is accessible from the "SOFTWARE" link on the PDB home page and at http://www.rcsb.org/pdb/software-list.html. Requests to add macromolecular-related software links to this page may be sent to [email protected].
13-Aug-2002
PDB at the Protein Society Symposium
The PDB will host an exhibit booth at the 16th Annual Symposium of the Protein Society, to be held at the San Diego Marriott Hotel and Marina on August 18-20, 2002. We look forward to meeting many of our users at this event - please stop by and visit the PDB team members in booth 204.
PDB CD-ROM Set #101 Released
The current Protein Data Bank CD-ROM set (release #101) is now being distributed. This release contains the macromolecular structure entries for the 18,528 structures available on the PDB Web site as of July 1, 2002.
The CD-ROMs are produced quarterly as of the last update of the PDB Web site for March, June, September and December. Further information is available at http://www.rcsb.org/pdb/cdrom.html where the CD-ROM documentation can also be accessed.
06-Aug-2002
August 10: PDB Users Lunch at IUCr
The PDB's Users Lunch will be held on Saturday August 10, 2002, at 12:30 p.m. The lunch will take place in the Le Jura Restaurant of the PalExpo Center as part of the IUCr Congress and General Assembly in Geneva, Switzerland. Food and conversation about the PDB will be provided. We hope to see you there!
We would like to thank the companies that helped to support this event -- Area Detector Systems Corporation, Blake Industries, Inc., deCODE genetics Emerald BioStructures Products, GlaxoSmithKline, Hampton Research, IBM, Merck Co., Inc. - USA, and Schering-Plough Pharmaceuticals.
30-Jul-2002
PDB at ISMB and IUCr
Come join in on PDB-related activities at the ISMB and IUCr meetings!
The PDB will participate in the exhibition at the Tenth International Conference on Intelligent Systems for Molecular Biology (ISMB), to be held at the Shaw Conference Center in Edmonton, Alberta, Canada, on August 3-7. At booth #36, members of the PDB will be available to answer your questions.
The PDB will also particpate in the the XIX Congress of the International Union of Crystallography (IUCr), which will be held in Geneva, Switzerland, on August 6-15. PDB members will be exhibiting at booth #13, a users lunch will be held on August 10th in Le Jura, and a session on databases will be held on August 13th.
We hope to see you at these events!
23-Jul-2002
Structural Genomics Informatics and Software Integration Workshop Web Site Available
The Structural Genomics Informatics and Software Integration (SG ISI) workshop was held at the Hyatt Regency Hotel in San Antonio on May 24-25, 2002. This workshop organized by Helen Berman, Tom Terwilliger and John Westbrook was attended by more than forty people including software developers, people involved in data management for structural genomics projects, and representatives of the Protein Data Bank (PDB). The workshop focused on data specification, software integration, and data management issues associated with automated deposition of data into the PDB from high-throughput structure determinations.
The materials from this meeting have been made available at http://deposit.pdb.org/sgisi02/.
16-Jul-2002
New Features on the PDB Web Site
Three new features are now available from the primary PDB Web site and its mirror sites:
- After a period of testing on the PDB beta Web site, the Swiss-Pdb Viewer is now linked from the View Structure section of the
Structure Explorer page for each PDB entry.
- The Citation Tabular Reports, which can provide bibliographic information for each structure in a search results set, now include links to search Medline either by PDB ID or Medline ID.
- A Perl script to download PDB files from any given update date is now available from ftp://ftp.rcsb.org/pub/pdb/software/. This script has three usages:
getPdbUpdate.pl dates -- retrieves and prints a list of valid update dates;
getPdbUpdate.pl latest -- retrieves all files from the latest update; and
getPdbUpdate.pl 20020603 -- retrieves all files from that particular update.
Questions or comments on these new features may be sent to [email protected].
09-Jul-2002
PDB Newsletter 14 Now Available
The Summer 2002 issue of the PDB Newsletter has been released in HTML format at http://www.rcsb.org/pdb/newsletter/2002q2/. This document describes PDB's endeavors and developments over the past three months in the areas of data deposition and processing; data query, reporting, and access; and outreach. This issue will soon be available in .pdf and print format as well. Subscriptions for the quarterly printed distribution of the PDB newsletter may be requested by sending your postal address to [email protected].
02-Jul-2002
Target Registration Database Available for Structure Genomics Projects Worldwide
TargetDB is a target registration database that was originally developed to provide registration and tracking information for NIH P50 structural genomics centers. TargetDB has now been expanded to include target data from worldwide structural genomics and proteomics projects. The scope of TargetDB is to provide timely status and tracking information on the progress of the production and solution of structures.
The target database can be searched by sequence using FASTA (Pearson, W.R. and Lipman, D.J. (1988) "Improved tools for biological sequence comparison" PNAS 85:2444-2448). Sequence searches may include only the target sequences or the PDB sequences. Target sequences may also be searched by contributing site, protein name, project tracking identifier, date of last modification, and the current status of the target (e.g. cloned, expressed, crystallized, ...). Search results may be viewed as HTML reports, FASTA data files, or in XML.
Information on target classification is also available from http://presage.berkeley.edu and http://proteome.umbi.umd.edu.

25-Jun-2002
PDB Paper Published in American Scientist
As highlighted on its cover, the July-August issue 2002 of the American Scientist contains an article that explores the history of protein structure determination and the PDB. "Protein Structures: From Famine to Feast" also reports how structural genomics initiatives will populate the PDB with many unique structures in the future. The illustrations were created by David S. Goodsell, author and illustrator of the PDB's Molecule of the Month feature and Molecular Machinery poster.
Protein Structures: From Famine to Feast
Helen M. Berman, David S. Goodsell and Philip E. Bourne
American Scientist (2002) 90:4, pp. 350 - 359.
18-Jun-2002
Theoretical Model Files Soon to be Moved to Separate Directory on PDB FTP Site
As previously announced, the PDB will separate theoretical model coordinate files from the main archive beginning July 1, 2002. After this date, the main archive will consist of structures determined using experimental methods only. Theoretical models will only be available for download from the PDB FTP site as follows:
- All theoretical models will be moved into a separate location in the FTP archive, and a subdirectory for obsoleted models
will also become available:
pub/pdb/data/structures/models/current
Any newly deposited or released theoretical model will go directly into the /models/current directory. If it supersedes an earlier model, that earlier model will be obsoleted, and moved from /models/current to /models/obsolete.
pub/pdb/data/structures/models/obsolete - Index files will be available in the models directories to facilitate browsing.
- From the Web interfaces, a theoretical model file will only be accessible by entering its PDB ID. All other queries (SearchLite, SearchFields, Status) will not return model entries.
- Searches by the PDB ID of a model through the Web will return a hyperlink to that model's coordinate file in the FTP archive.
- Theoretical models will be removed from the PDB Contents Growth and PDB Holdings statistics at http://www.rcsb.org/pdb/holdings.html.
Theoretical models will also be moved to a separate location on the PDB CD-ROMs after July 1.
Models may still be deposited after this date, using ADIT: http://pdb.rutgers.edu/adit/, http://pdbdep.protein.osaka-u.ac.jp/adit/; or AutoDep: http://autodep.ebi.ac.uk/. These depositions will be forwarded to the models directory of the PDB FTP server without further annotation or validation.
11-Jun-2002
Two PDB Papers Published
Two papers on the PDB resource have recently been published in a special issue of Acta Crystallographica on crystallographic databases.
A paper entitled, "The Protein Data Bank", was featured in this issue. This paper introduced and described the goals of the PDB, the systems in place for data deposition and access, and plans for the future development of the resource.
The article, "Protein structure resources", was also included in this periodical. In this article, web-accessible resources derived from data in the PDB were classified and described. These include resources for protein structure and functional classification, as well as links to primary genomic information, protein-protein interactions, protein dynamics, and protein-modeling resources.
Acta Crystallographica Section D: Biological Crystallography
Volume 58, Part 6 Number 1 (June 2002)
Copyright (c) International Union of Crystallography 2002
The Protein Data Bank.
H.M. Berman, T. Battistuz, T.N. Bhat, W.F. Bluhm, P.E. Bourne, K.
Burkhardt, Z. Feng, G.L. Gilliland, L. Iype, S. Jain, P. Fagan, J.
Marvin, D. Padilla, V. Ravichandran, B. Schneider, N. Thanki, H.
Weissig, J.D. Westbrook and C. Zardecki.
Acta Cryst. (2002). D58, pp. 899-907.
Protein structure resources.
H. Weissig and P.E. Bourne.
Acta Cryst. (2002). D58, pp. 908-915.
4-Jun-2002
New PDB Features
The Swiss-PdbViewer and new external links are now available from the Structure Explorer page for each PDB entry.
The Swiss-PdbViewer is now included as an option in the View Structure section of Structure Explorer pages on the PDB beta Web site. This molecular graphics viewing program allows users to load and display several molecules simultaneously, create layered image files, and more. Instructions for downloading and configuring the Swiss-PdbViewer can be found at http://www.rcsb.org/pdb/help-graphics.html#spdbv_download.
New external links to PDBsum, CATH, and SCOP are now accessible from the Summary Information section of the Structure Explorer pages on the PDB Web site and its mirrors. Additionally, external links to analysis resources, such as Contacts of Structural Units, are now available from the Geometry section of the Structure Explorer pages on these sites.
Questions about these new features may be sent to [email protected].
28-May-2002
PDB CD-ROM Set #100 Now Available
The current Protein Data Bank CD-ROM set (release #100) is now being distributed. This release contains the macromolecular structure entries for the 17,679 structures available on the PDB website as of April 1, 2002.
The CD-ROMs are produced quarterly as of the last update of the PDB Web site for March, June, September and December. Further information is available at http://www.rcsb.org/pdb/cdrom.html where the CD-ROM documentation can also be accessed.
21-May-2002
PDB at the ACA and the International School of Crystallography
The PDB will participate in two upcoming meetings this month. We are looking forward to seeing everyone at the American Crystallographic Association's Annual Meeting in San Antonio, Texas (May 25-30). The PDB will be exhibiting at Booth #413.
On Friday, May 24, a PDB talk will be given as part of the International School of Crystallography Meeting in Erice, Italy. We hope you can join us at these events!
14-May-2002
Deposition Checklists Available
Checklists of the data items to have on hand when depositing structures via ADIT are available for both X-ray and NMR depositions. These checklists highlight the information that that will be requested when depositing.
ADIT is accessible from http://deposit.pdb.org/adit/ (RCSB-US) and http://pdbdep.protein.osaka-u.ac.jp/adit/ (Osaka University, Japan). Further information about PDB depositions is available at http://deposit.pdb.org/.
7-May-2002
PDB Focus: Query Result Browser Options
Any set of structures returned from a PDB search can be included in a tabular report, downloaded in one file, queried further in a refined search, and more when using the Query Result Browser.
The pull down menu at the top of the Query Result Browser page can be used to:
- Perform a New Search -- returns either SearchFields or SearchLite, depending on which was used for the previous search.
- Download Structures or Sequences -- search results can be downloaded in a single file in a variety of file and compression formats
- Refine Your Query -- the resulting set can be searched further with the option to remove files that contain certain parameters or keywords
- Create a Tabular Report -- choose from a variety of prepared reports (Structure Summary, Sequence, Crystallization Description, Unit Cell, Data Collection, Refinement, Citation) or customize your own
- Select/Deselect All Structures -- all the entries can be selected/deselected with one click. This is useful when performing further operations on a result set in which the majority, but not all, structures are desired.
- Remove Sequence Homologues -- creates a subset of the structures from which sequence homologues have been largely removed
- Show Only Selected Structures -- removes unselected structures from the browser view
- Show Structures on Hold -- returns a list of unreleased structures which match the query.
- Review Your Query -- shows the search parameters used and the number of structures found, selected, and on hold.
These options are activated by selecting one and clicking on the "Go" button.
More information on using the options available from the Query Result Browser is available at http://www.rcsb.org/pdb/help-results.html.
30-Apr-2002
PDB Focus: Restarting ADIT depositions
Depositing a structure using ADIT can be done in more than one session by using the "Session Restart ID". This identifier appears in red in the center of the browser window when the ADIT "deposit" step is first started. It also appears in the title of the browser throughout the deposition session.
ADIT stores the data entered in a category every time the user presses the SAVE button. These data will be available with the restart ID until the user deposits the entry by selecting the "DEPOSIT NOW" button.
The restart ID is entered in the space provided on the ADIT home page to return to the undeposited ADIT entry. Restart IDs are case-sensitive.
ADIT is available at http://deposit.pdb.org/adit/ (RCSB-US) and http://pdbdep.protein.osaka-u.ac.jp/adit/ (Osaka University, Japan).
A tutorial guide to using ADIT is available in English at http://pdb.rutgers.edu/adit/docs/tutorial.html and in Japanese at http://www.protein.osaka-u.ac.jp/pdb/. Example "in progress" ADIT depositions are available at http://pdb.rutgers.edu:81/.
23-Apr-2002
PDB Focus: Software Page
The PDB's Software page is a portal to software developed by the RCSB and others in the macromolecular structure community. RCSB-developed software, such as the CIFTr application for translating files between mmCIF and PDB formats, the STAR (CIF) modules for parsing mmCIF files, and the ADIT workstation version are available for download. Links to external software resources relating to mmCIF, crystallography, NMR, structure analysis and verification, modeling and simulation, and molecular graphics are also available here.
This page is accessible from the "SOFTWARE" link on the PDB home page and at http://www.rcsb.org/pdb/software-list.html. Requests to add macromolecular-related software links to this page may be sent to [email protected].
16-Apr-2002
BioSync: A Structural Biologist's Guide to Synchrotron Facilities
The Structural Biology Synchrotron Users Organization (BioSync) offers a portal to resources for crystallographers through the BioSync Web site at http://biosync.sdsc.edu/. This site contains information for prospective synchrotron users in the field of macromolecular crystallography, including technical desciptions of U.S. beamlines. Hyperlinks to beam time request forms, synchrotron usage training, site contact information and directions are also available. Further details can be obtained by visiting the BioSync Web site or by sending email to [email protected]. Further information on BioSync can also be found in the article:
A biologist's guide to synchrotron facilities: the BioSync web resource
Anne Kuller, Ward Fleri, Wolfgang F. Bluhm, Janet L. Smith, John Westbrook and Philip E. Bourne
Trends in Biochemical Sciences 27:4, pp. 213-215
(1 April 2002)
9-Apr-2002
PDB Newsletter 13 Released
The Spring edition of the Protein Data Bank Newsletter is now available in HTML format at http://www.rcsb.org/pdb/newsletter/2002q1/. This document outlines the latest activities of the PDB in the areas of data deposition and processing; data query, access and reporting; and outreach. A plain text version can be accessed at ftp://ftp.rcsb.org/pub/pdb/doc/newsletters/rcsb/. This issue will soon be available in .pdf and print format as well. Subscriptions for the quarterly printed distribution of the PDB newsletter may be requested at [email protected].
2-Apr-2002
Theoretical Model Coordinate Files to be Moved to Separate PDB FTP Directory
After much discussion with the user community over the last several years, and upon the recommendation of the PDB Advisory Committee, the PDB will separate theoretical models from the main archive. Beginning July 1, 2002, the main archive will consist of structures determined using experimental methods only. Theoretical models will be available for download from the PDB FTP site, and pointers to this location will be available from user query result summaries that include models in the result set. Theoretical models will also be moved to a separate location on the PDB CD-ROMs after July 1. Further details regarding the retrieval of models will be disseminated in the near future.
Models may still be deposited after this date using ADIT: http://pdb.rutgers.edu/adit/, http://pdbdep.protein.osaka-u.ac.jp/adit/; or AutoDep: http://autodep.ebi.ac.uk/. These depositions will be forwarded to the models directory of the PDB FTP server without further annotation or validation.
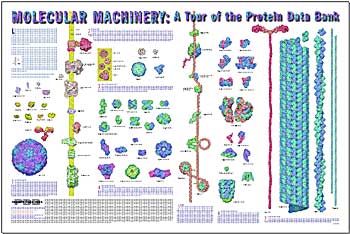 26-Mar-2002
26-Mar-2002
PDB Molecular Machinery Poster Released
The PDB is pleased to announce the release of a poster entitled, "Molecular Machinery: A Tour of the Protein Data Bank". This poster features illustrations of 75 select structures from the PDB, showing their relative sizes at a scale of three million to one, and generally describes their critical roles in the functions of living cells. Content for the poster was provided by David Goodsell of The Scripps Research Institute, with graphic design by Gail Bamber of the San Diego Supercomputer Center. Copies of this poster can be obtained by sending your request to [email protected].
19-Mar-2002
Experimental Data Availability Included in Status Search Results
A search of unreleased structures now indicates if the structure factor or constraint file has been deposited, and when this file will be released. This feature was added to the primary PDB Web site and its mirrors after a period of testing on the beta web site. Feedback on this new feature may be sent to [email protected].
12-Mar-2002
PDB Focus: Annotating Data Around the World
PDB data is processed by an international effort. ADIT deposition sites are located at the RCSB-Rutgers site (US) and at the Institute for Protein Research, Osaka University (Japan).
Structures deposited using ADIT are processed immediately and returned to the author. The files are fully processed and are released according to the release status provided by the author. Data are processed by staff from the RCSB (at Rutgers University in New Jersey and remotely at the Center for Complex Molecular Systems and Biomolecules in the Czech Republic) and from the Institute for Protein Research at Osaka University. The procedures used for processing these data are described in the PDB Data Processing and Annotation Procedures at http://www.rcsb.org/pdb/info.html#File_Formats_and_Standards.
Structures are also deposited using AutoDep at the European Bioinformatics Institute (EBI) in the United Kingdom. Data deposited using AutoDep are processed by the EBI.
|
ADIT
|
RCSB Osaka University |
http://pdb.rutgers.edu/adit/ http://pdbdep.protein.osaka-u.ac.jp/adit/ |
|
AutoDep
|
EBI | http://autodep.ebi.ac.uk/ |
5-Mar-2002
PDB CD-ROM Issue 99 Released
The latest PDB CD-ROM (release #99) set is currently being distributed. This release contains the macromolecular structure entries for the 16,972 structures available as of January 1, 2002. The CD-ROMs are produced quarterly as of the last update of the PDB Web site for March, June, September and December. The experimental data (X-ray structure factors and NMR constraints) are also included, if available. Further information is available at http://www.rcsb.org/pdb/cdrom.html where the CD-ROM documentation can also be accessed.
26-Feb-2002
Protein Art Proves to be Popular
 |
| "The Art of Science" opening reception at The Gallery |
Many students, professors, and local educators viewed the exhibit during its run. Many commented on how they were surprised by how beautiful they found the images, and how interested they were in the scientific descriptions that accompanied the pictures. The exhibit also was the focus of several newspaper articles, including a story in The Star-Ledger and a front page article in The Bergen Record.
Various representations of proteins found in the PDB were highlighted, including large scale depictions of the images available from PDB Structure Explorer pages, images of collagen by Jordi Bella, and pictures from the PDB's Molecule of the Month series by David S. Goodsell. The exhibit, which was open from January 21 - February 9, was curated by Christine Zardecki.
19-Feb-2002
Experimental Data Availability in Status Reports on PDB Beta Web Site
A new field has been added to the Status Search results on the PDB beta Web site at http://beta.rcsb.org/pdb/status.html. The "Exp. Data" line now informs users whether or not structure factors or NMR constraints have been deposited with the coordinates of a structure. For entries that include either structure factors or NMR constraints, it also notes when this experimental data will be released. Comments on this enhancement may be sent to [email protected].
PDB Newsletter 12 Released
Issue 12 of the PDB newsletter is now available in HTML format at http://www.rcsb.org/pdb/newsletter/2001q4/. This document details recent activities of the PDB from the last quarter of 2001, including the latest developments in data deposition and processing; data query, access and reporting; and outreach. A plain text version can be accessed at ftp://ftp.rcsb.org/pub/pdb/doc/newsletters/rcsb/. This issue will soon be available in .pdf and print format as well. Subscriptions for the quarterly printed distribution of the PDB newsletter may be requested at [email protected].
12-Feb-2002
PDB at the Biophysical Society Meeting
The PDB will participate in the exhibition at the 46th Annual Meeting of the Biophysical Society, to be held on February 23-27, 2002, at the Moscone Convention Center in San Francisco, CA. PDB staff will be available at booth #204 to answer questions. We hope to see you there!
 |
|
Click on the image to enlarge
|
5-Feb-2002
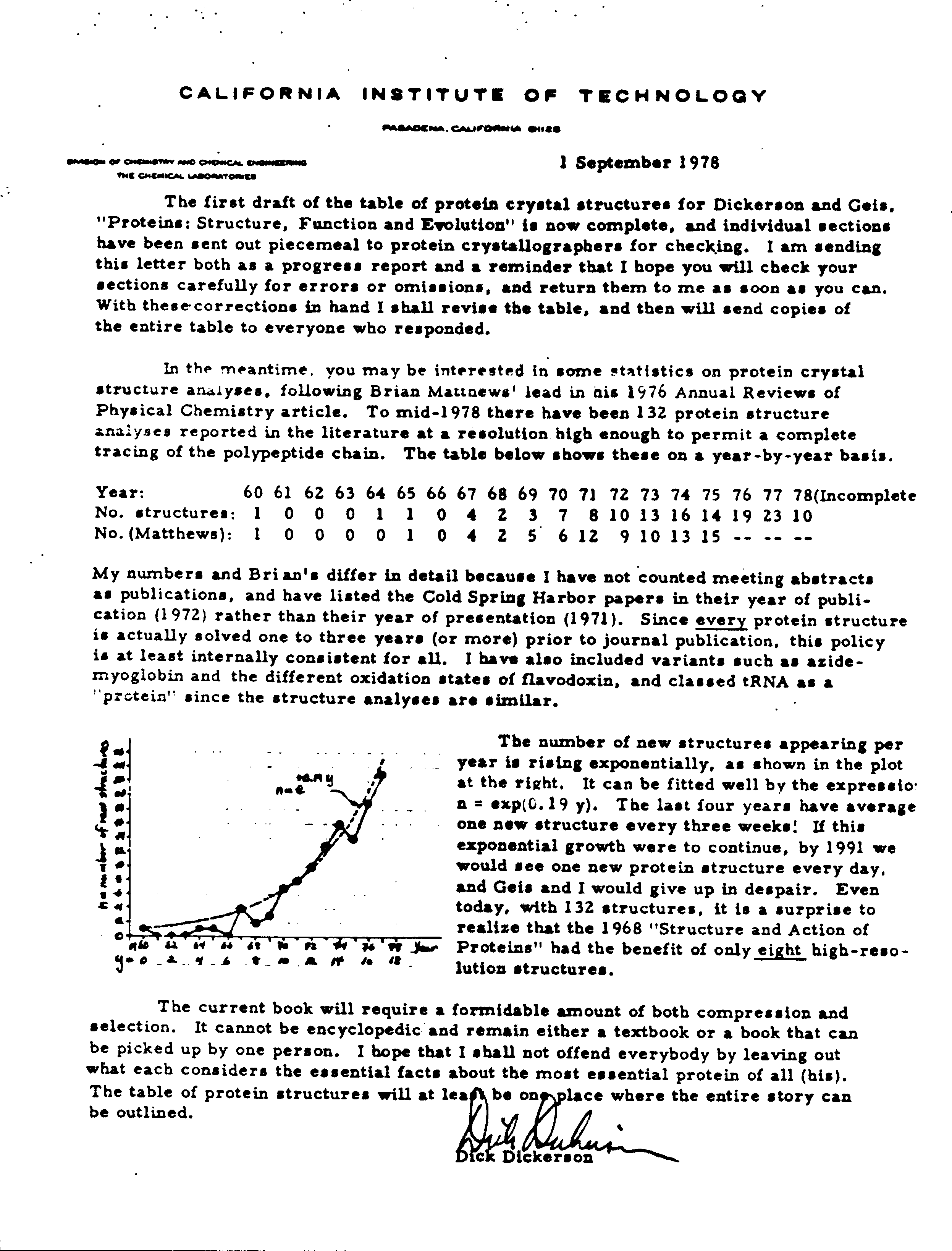
Rate of PDB Holdings Growth Predicted in 1978?
In 1978, Richard E. Dickerson examined the number of available crystal structures. Based upon that number, he came upon an equation to describe the exponential growth for solved crystal structures.
In a letter describing a book he was working on with Irving Geis, Dickerson noted (and illustrated with a hand-drawn graph) that the number of new structures appeared to be following the exponential law n = exp(0.19 y), where n is the number of new structures per year and y is the year number since 1960. This equation predicted that at the end of 2001, there would be 13,941 crystal structure entries available in the PDB (14,000 crystal structures are currently available). Using this equation, there should be 24667 crystal structures in 2004.
Thanks to Arthur Arnone, who noticed that there were 57 structures more than Dickerson's equation predicted in March 2001. Predictions for the rate of NMR structures may be sent to [email protected].
29-Jan-2002
Phase Out of BNL FTP Archive Reminder
Since 1999, the Research Collaboratory for Structural Bioinformatics (RCSB) has maintained two distinct FTP sites: the RCSB Protein Data Bank (PDB) site at ftp://ftp.rcsb.org/ (and its mirrors; see http://www.rcsb.org/pdb/mirrors.html), and the Brookhaven National Laboratory (BNL) PDB site at ftp://bnlarchive.rcsb.org.
In order to conserve resources and avoid confusion arising through the existence of two distinct PDB FTP sites, the RCSB will phase out the BNL PDB archive as of March 1, 2002, as announced on October 19, 2001 (http://www.rcsb.org/pdb/lists/pdb-l/200110/msg00024.html). This decision was made after consultation with the PDB Advisory Committee and review by members of the PDB user community.
The files previously available only at ftp://bnlarchive.rcsb.org/pub/resources/index/ are now available at ftp://ftp.rcsb.org/pub/pdb/derived_data/index/.
Current users of the BNL PDB archive are encouraged to consider the option of mirroring the RCSB FTP archive. The RCSB FTP archive can be found at ftp://ftp.rcsb.org and instructions for mirroring it can be found at http://www.rcsb.org/pdb/ftpproc.final.html.
A Perl script is provided to assist with conversion of existing BNL FTP directory structure to the RCSB FTP directory structure. Further information about the script is available at ftp://ftp.rcsb.org/pub/pdb/software/bnl2rcsb.pl.
Please send your questions or concerns, or requests for assistance regarding this change, to [email protected].
22-Jan-2002
STING Millennium Suite Released on PDB Web Site
After a period of testing at the PDB beta test site, select components of STING Millennium Suite (SMS) are now available from the Structure Explorer pages of the primary PDB Web site and its mirrors. SMS, a set of Java-based tools for the simultaneous display of information about macromolecular structure and sequence, was developed by Dr. Goran Neshich of Embrapa-CNPTIA (Campinas, Brazil) and colleagues, in collaboration with Dr. Barry Honig's laboratory at Columbia University in New York City, NY. The SMS links from the PDB site are served by an SMS mirror that is being maintained at the San Diego Supercomputer Center.
The "Sequence Details" and "View Structure" sections of the Structure Explorer now link to two interactive structure and sequence SMS views for any PDB structure, which include options to access features such as a graphical display of amino acid contacts; these views require Chime and a Java-enabled Web browser. A simpler "Protein Dossier" view is also available from the "Sequence Details" section, offering a static graphical summary of sequence-and structure-based properties, such as relative entropy and temperature factors.
The "Geometry" section of the Structure Explorer now links to a Ramachandran plot for each PDB entry, also served from SMS, with options including the inter-connection of data in a dihedral angle plot with the 3-D structure of the molecule. This view also requires Java and Chime. SMS is also accessible from the "Other Sources" section of the Structure Explorer for each PDB entry, under the category of Visualization resources.
Further information about this suite of tools is available from the SMS home page at http://mirrors.rcsb.org/SMS/, and at http://www.rcsb.org/pdb/help-results.html. Comments may be sent to [email protected].
"The Art of Science" -- A PDB Art Gallery Exhibit
Images from the Protein Data Bank are being presented as "The Art of Science", an art exhibit appearing at Rutgers University that looks at the beauty inherent in the three-dimensional structures of proteins.
Various representations of proteins found in the PDB are highlighted, including large scale depictions of the images available from PDB Structure Explorer pages, images of collagen by Jordi Bella, and pictures from the PDB's Molecule of the Month series by David S. Goodsell. The exhibit was curated by Christine Zardecki.
"The Art of Science" is on display at The Gallery, a space dedicated to art exhibits at Rutgers University. The Gallery is located in the Busch Campus Center (604 Bartholomew Road, Piscataway, NJ). "The Art of Science" will be on display from January 21 - February 9, 2002 (Monday - Friday, 11AM to 10PM and Saturday - Sunday, 12PM to 4PM).
15-Jan-2002
PDB Focus: Pretest New Query Features at the PDB Beta Web Site
New query features, such as the recent additions of the STING Millennium Suite to Structure Explorer pages and the ability to search on a subset of non-homologous structures, are always made available for public testing at the PDB Beta Web Site at http://beta.rcsb.org/pdb/.
New developments ready for testing are announced on these pages. After the new features have been reviewed by the public, they are incorporated into all of the PDB sites. Comments about features being tested at the PDB Beta Test Site should be sent to [email protected]. We thank all of you who have used this site and have provided feedback!
8-Jan-2002
Data Uniformity Paper Published
The latest Nucleic Acids Research issue features a paper from the PDB entitled "The Protein Data Bank: unifying the archive", that describes the ongoing efforts of the data uniformity project which addresses inconsistencies in the archive.
Updates on the Data Uniformity Project are posted at http://www.rcsb.org/pdb/uniformity/index.html.
Nucleic Acids Research, 2002, Vol. 30, No. 1 245-248
© 2002 Oxford University Press
The Protein Data Bank: unifying the archive
John Westbrook, Zukang Feng, Shri Jain, T. N. Bhat, Narmada Thanki, Veerasamy Ravichandran, Gary L. Gilliland, Wolfgang Bluhm, Helge Weissig, Douglas S. Greer, Philip E. Bourne and Helen M. Berman









 26-Mar-2002
26-Mar-2002




