RCSB PDB Help
Sequence Viewers
Sequence Annotations Viewer
The Sequence Annotations Viewer provides graphical summaries of PDB protein biological and structural features and their relationships with UniProtKB entries. Protein alignments and features are loaded from RCSB PDB web services including the main data API and the 1D Coordinate Server. These resources integrate information from multiple resources and databases. Structural features such as secondary structure, angle/distance outliers, protein-ligand binding sites or disulfide bridges, are extracted from the PDB structural data. Additionally, structural domains are annotated from CATH and SCOPe databases. Biochemical and biomedical features are collected from the UniProtKB database and mapped onto PDB sequences.
Responsive and Interactive Properties
- Zoom using the mouse scroll (wheel) or the mousepad/touchpad to see more details and to show the sequence of amino acids
- To zoom using the mousepad/touchpad hover over the Sequence Annotations Viewer, and move/drag 2 fingers in an up and down motion
- Once zoomed, left click and drag to move the panel left or right (using either a mouse or touchpad)
- Left click on a feature to highlight the annotated region or right click to select and highlight a custom area
- Hovering over track features will display a legend at the top right corner of the viewer (see Feature Tooltips)
Conventions
- The first track defines the sequence reference over which features are mapped
- A white circle over the start (or end) of a block indicates that the feature in its original reference does not start (or end) in that position but before (or after)
- A dashed line between blocks indicates that in the original reference the feature is connected and therefore a sequence insertion has occurred
- Color flags (blue/orange) between track titles and track features indicates feature provenance (blue for RCSB PDB and orange for third party resources)
Structure Summary Page - Sequence Tab
In the Sequence Tab (e.g. 4z35) the Sequence Annotations Viewer will display the full range of available features (structural and biological annotations) and the alignments between Polymer Instances (chains) and UniProtKB sequences. The select menu can be used to display the features for the different PDB Polymer Instances (chains) of the current Entry (Figure 1).
Structure Summary Page - Main Tab
The Sequence Annotations Viewer in the Structure Summary Pages (e.g. 4z35) displays the alignments between PDB Entity sequences and UniProtKB. Additionally, it includes Entity features such as mutations and artifacts, and the different regions that has not been (or partially) observed at Instance level. The select menu can be used to change the reference (Entity/UniProtKB) over which features are mapped and displayed (Figure 2).
Disorder and disorded binding values are calculated based on sequence alone using IUPred (Dosztányi Z., 2018) and ANCHOR (Dosztányi, Z., et al., 2009) algorithms, respectively. The reported disorder values for each residue range between 0 and 1, corresponding to its probability of being part of a disordered region. The lower the probability, the less likely a given residue is within disorder region. The 0.5 cutoff is used for coloring, blue more likely to be an ordered region, red - disordered.
Hydropathy is calculated based on sequence alone using the Kyte-Doolittle hydrophobicity scale (Kyte and Doolittle, 1982).
Feature Tooltips
Feature tooltips contain important information about feature positions in the current and original reference sequence. When the cursor is hovering over specific amino acid or sequence features, relevant tooltips are displayed top right corner of the Sequence Annotations Viewer (Figure 3)
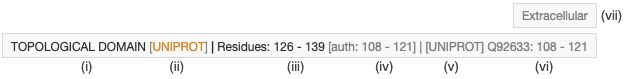
|
| Figure 3: Tooltips that appear when the cursor is hovering over a specific feature in the Sequence Annotations Viewer |
- (i) Feature short name description
- (ii) Database source or software name used to obtain the feature. Color indicates its provenance (blue RCSB PDB and orange third party resources)
- (iii) Feature position in the current reference system
- (iv) PDB residue author Ids associated to the feature position
- (v) Feature source database/software name
- (vi) Feature original reference entry Id and positions
- (vii) Feature description
For Developers
The Sequence Annotations Viewer (previously called Protein Feature View) is an Open Source project written in TypeScript. It is available at github.com














