RCSB PDB Help
Sequence Viewers
Genome View
Overview
The Genome View provides graphical summaries of the correspondences between PDB entity sequences and genomes. Protein and genome alignments are loaded from the RCSB PDB 1D Coordinate Server. This resource integrates alignments from multiple resources and databases including UniProtKB, RefSeq and SIFTS. This view will display what regions of the different gene products are covered by the PDB entity and will allow to explore the relationships between residue indexes and their corresponding nucleotide coordinates at genome level.
Genome Viewer Use
The Genome Viewer can be used:
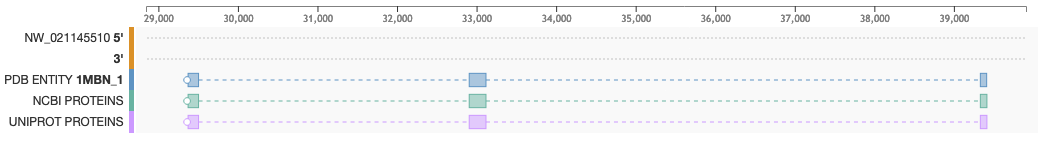
- To see the mapping between proteins and genes: Blocks display the position of PDB Entities (Blue), NCBI proteins (Green) and UniProt proteins (purple)
- To see the genome reading frame: A white circle points to the reading frame direction. If the circle is located on the left side of the track, the protein is mapped onto the negative DNA strand (3') and the sequence is read right to left. If the circle is on the right side, the protein is mapped on the positive strand (5') and is read left to right
- To see intron/exon boundaries: Exons are represented as solid boxes while introns are displayed as dashed lines connecting boxes
Documentation
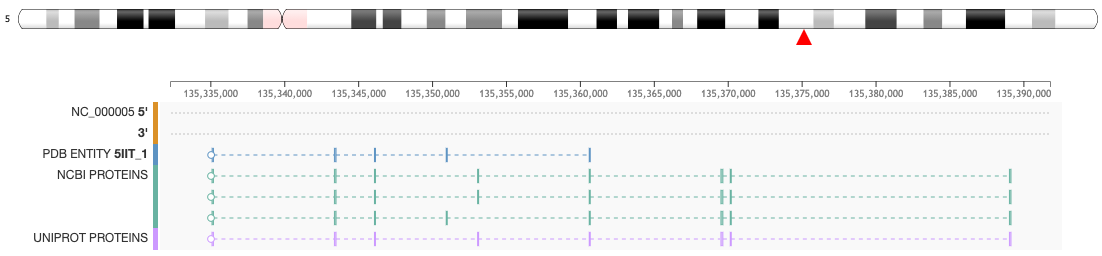
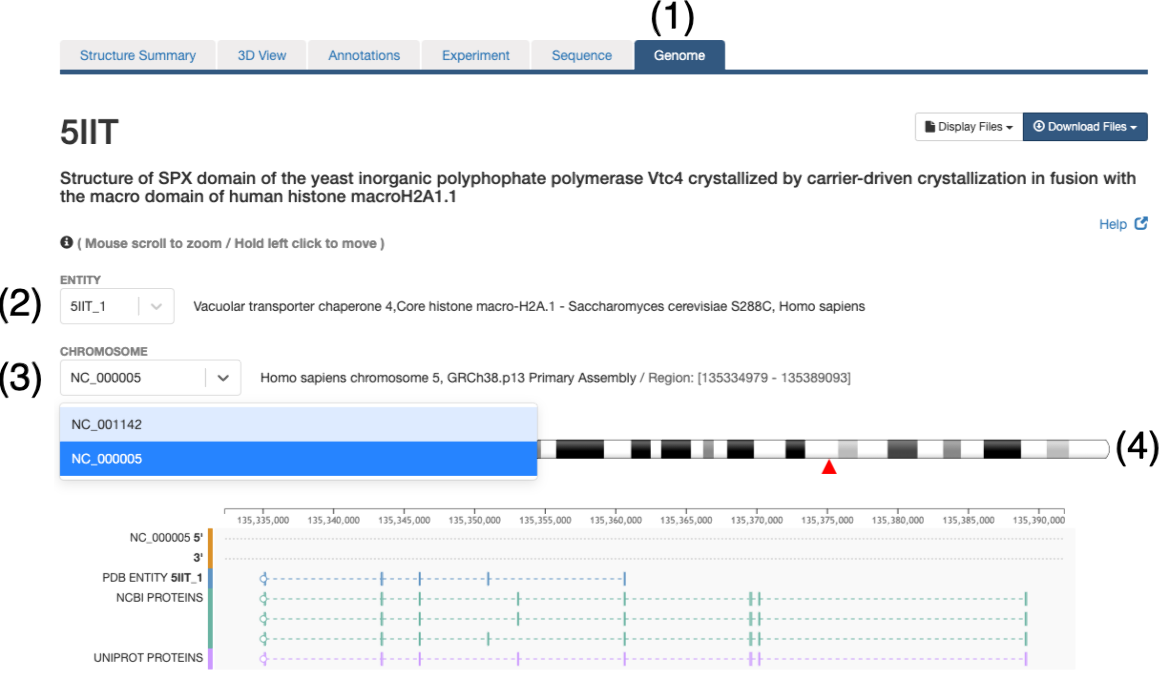
This feature is available from the Genome View Tab on the Structure Summary Page. For example, clicking on the Genome Tab for a PDB entry (e.g. 5IIT) the mapping of the protein sequence of the protein entities (UniProtKB and NCBI RefSeq) is displayed at genome level.
For each mapping the following conventions are used:
- The first pair of tracks display the DNA sequences 5' and 3', respectively, over which PDB entity and proteins are mapped
- Proteins are represented as boxes connected by dashed lines. Connected boxes correspond to the codifying regions of a gene
- The protein sequence of the selected PDB entity is shown in blue boxes connected by dashed lines
- The bottom two tracks list the protein sequence from NCBI RefSeq (green flag tracks) and UniProtKB (purple flag tracks)
- A white circle on the left or right of a protein track points to the reading frame direction of the gene. For instance, if the circle is on the left, the protein sequence is to be read right-to-left and corresponds to the negative (3') strand
- When the chromosome ideogram is available, a red triangle will indicate the position of the PDB entity at chromosome level
How-To
- In order to explore the protein-genome mapping click on the Genome Tab of the Structure Summary Page (e.g. 5IIT)
- In this page a drop down menu will allow selection of the entity to explore. Only those entities that could be mapped to a reference genome will be available
- A particular entity may be composed of different protein fragments and thus, it may map to multiple chromosomes, the second drop down menu will allow to explore the different chromosomes
- Commonly a chromosome ideogram will be displayed and the entity position will be indicated with a red triangle
Responsive and Interactive Properties
- Zoom using the mouse scroll (wheel) or the mousepad/touchpad to see more details and to show the sequence amino and nucleic acids
- To zoom using the mousepad/touchpad hover over the Protein Feature View, and move/drag 2 fingers in an up and down motion
- Once zoomed, left click and drag to move the panel left or right (using either a mouse or touchpad)
For Advanced Users
Genome / Structure alignments API is available at 1D Coordinate Server












