2003 PDB News
Contents:
Read the latest PDB news. Earlier news is available and is archived in the RCSB PDB newsletters.
30-Dec-2003
 |
|
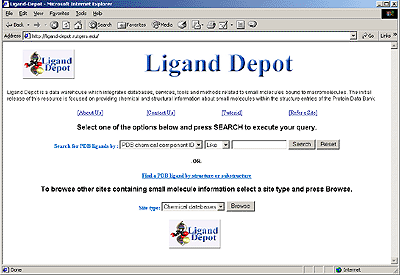
The Ligand Depot Interface
|
Ligand Depot--a Small Molecule Information Resource
Ligand Depot (http://ligand-depot.rutgers.edu) is a data warehouse that integrates databases, services, and tools related to small molecules bound to macromolecules. The initial release (v. 1.0, November, 2003) focuses on providing chemical and structural information for ligands that are found as part of the structures deposited with the PDB.
Ligand Depot allows users to extract ligand information from the PDB, to perform chemical substructure searches, and to search other small molecule resources on the Web. One of the distinguishing features of Ligand Depot is that it allows users to retrieve the coordinates of any small molecule found within the structure entries of the PDB. It is also updated daily and therefore provides the most current information on small molecules present in the PDB.
Ligand Depot currently includes chemical descriptions for the ~4,600 ligands that are part of the structures deposited in the PDB, and it offers various search options for obtaining information on these small molecules. It accepts keyword queries based on PDB ligand code, compound name and chemical formula. Using a simple graphical interface, a substructure search may also be performed between a small molecule of interest and all of the ligands present in the PDB.
Ligand Depot can also be used to browse a variety of other small molecule resources on the Web. Information from 70 small molecule sites are stored in Ligand Depot. These resources are organized into four categories, including molecular visualization sites, commercial sites, nomenclature sites, and chemical databases. Keyword searches may be performed on these external Web sites if they are search-enabled. Thus, information on ligands can be extracted from a diverse collection of Web resources using a single query.
A helpful tutorial for using Ligand Depot is accessible at http://ligand-depot.rutgers.edu/html1/User_Guides.html.
Happy Holidays from the RCSB PDB
The RCSB PDB staff wish to extend our best wishes to the community for a happy holiday season and a wonderful new year!
23-Dec-2003
RCSB PDB Article Published in Nucleic Acids Research
The article, "The distribution and query systems of the RCSB Protein Data Bank," has been published in the latest issue of Nucleic Acids Research. This feature describes the dissemination and accessibility of PDB data via the current RCSB PDB query and distribution system. It also introduces an alpha version of the future re-engineered system that will be released in beta during the first quarter of 2004. The abstract and full text of the article are also available from the Nucleic Acids Research Web site.
P.E. Bourne, K.J. Addess, W.F. Bluhm, L. Chen, N. Deshpande, Z. Feng, W. Fleri, R. Green, J.C. Merino-Ott, W. Townsend-Merino, H. Weissig, J. Westbrook and H.M. Berman (2004): The distribution and query systems of the RCSB Protein Data Bank. Nucleic Acids Research 32, pp. D223-5.
16-Dec-2003
PDB Focus: Redundancy Reduction Cluster Data Available on the PDB FTP Site
The results of the weekly clustering of protein chains in the PDB are posted at ftp://ftp.rcsb.org/pub/pdb/derived_data/NR/. These clusters are used in the "remove sequence homologs" feature on SearchLite, SearchFields, and the PDB home page on the PDB web sites.
Files that list the clusters and their rankings at 50%, 70% and 90% sequence identity are available. Smaller rank numbers indicate higher (better) ranking. Chains with rank number 1 are ranked as the best representative of their cluster.
The contents of these files and the details of the clustering and ranking are further described at ftp://ftp.rcsb.org/pub/pdb/derived_data/NR/README and http://www.rcsb.org/pdb/redundancy.html.
9-Dec-2003
NIGMS News: PSI-2 and Structural Biology Roadmap RFA
Concept Clearance of the PSI-2 Production Phase
Plans for the next phase of the NIGMS Protein Structure Initiative (PSI) were announced at the recent NIGMS Council meeting (http://www.nigms.nih.gov/news/reports/council-psi-sept03.html). This phase will begin in 2005 with the grant announcement expected for early 2004. It is envisioned as an interacting network with large-scale research centers that will operate as high throughput structural genomics pipelines for protein production and structure determination. The plans approved by the Council also include the establishment of specialized research centers for development of new methods, technology, and approaches for the production and structure determination of especially challenging proteins, such as membrane proteins and proteins from humans and other higher eukaryotic organisms, as well as for projects to address technology barriers to high-throughput operation.
Since 2000, the NIGMS has funded nine pilot structural genomics research centers as part of its plan to reduce the costs and increase the success of the structural determination of proteins. The long- range goal of the PSI is to make the three-dimensional atomic-level structures of most proteins easily obtainable from knowledge of their corresponding DNA sequences. The pilot projects have focused on high throughput methods for structure determination in order to achieve these goals. For more information please visit http://www.nigms.nih.gov/psi.
Announcement of Structural Biology Roadmap RFA
Structural biology is also prominent in the plans of the NIH Roadmap for Medical Research (http://nihroadmap.nih.gov/structuralbiology/index.asp). The roadmap includes an RFA (request for applications) for Centers for Innovation in Membrane Protein Production. Letters of intent are due by February 5, 2004 with applications due by March 11, 2004.
2-Dec-2003
International Collaborators to Form the Worldwide Protein Data Bank
The Research Collaboratory for Structural Bioinformatics (RCSB), the Macromolecular Structure Database at the EMBL-European Bioinformatics Institute (MSD-EBI), and Protein Data Bank Japan (PDBj) have announced a collaboration to form the Worldwide Protein Data Bank (wwPDB; http://www.wwpdb.org/). The announcement is published in the December issue of Nature Structural Biology; a PDF version of the article is available.
The collaboration reflects the growing international and interdisciplinary nature of scientific research, and formalizes the global character of the PDB, which has been used as an international resource for the collection and sharing of three-dimensional information on proteins and other large molecules since its inception 32 years ago. The formation of the wwPDB will be transparent to users and will ensure the overall quality and consistency of data directly available through the PDB.
"By providing a formal mechanism for standardizing the presentation of PDB data, software developers and users of the data will be assured of consistent data. At the same time, it is hoped that this wwPDB will allow for individual creativity in how the data are presented and made available to the community," said Helen Berman, director of the RCSB PDB and Board of Governors Professor of Chemistry at Rutgers, The State University of New Jersey.
Kim Henrick, head of the MSD-EBI said, "The PDB is a canonical research resource that transcends both scientific and political boundaries. The wwPDB agreement among the three equal partners elevates the responsibility for the deposition and accessibility of the data to a global level. The EBI has been a longtime deposition site and advisor to the PDB and the evolution of that role is a welcome development."
Head of the PDBj group at the Institute for Protein Research in Osaka University, Haruki Nakamura said, "The PDBj has become the representative for the PDB throughout Asia and Oceania. With the recent explosion of interest in structural biology and bioinformatics research in the region, which would not be possible without the PDB, it is a natural step for us to formalize our involvement through the wwPDB."
The PDB is the single archive of biological macromolecular structure data, which is made freely and publicly available to researchers, educators, and students. Worldwide, the PDB receives over 60 million hits per year. As of October 28, 2003, it contained 22,984 structures, a number that has been growing exponentially.
According to a 10-year agreement signed by the 3 founding members of the wwPDB, the sites will share responsibilities in data deposition, data processing, and distribution. An international advisory board will be formed to support the collaboration.
18-Nov-2003
Updates of mmCIF Files on the PDB FTP Site
As previously announced, the update of September 2, 2003 included the replacement of all mmCIF files in the PDB FTP archives with the remediated mmCIF files. Due to our ongoing data curation efforts, occasional weekly updates will include the replacement of large numbers of mmCIF files. We have decided to reserve the first Tuesday of each month for these potential bulk mmCIF updates. Such updates should not be required every month. If you would like to be added to a list of FTP users who will receive individual e-mail notifications prior to each bulk update, please send your request to [email protected].
New Update Release of CD-ROM Sets
The October 2003 update of the PDB CD-ROM data set, Release #106, is an incremental set of 1,583 experimentally determined structures and 61 models. The structure coordinate files, contained on one disk, are shipping now.
Files for entries re-released for any reason between July and October 2003 are included in this update. A list of files that have become obsolete since the last update is included so users can update their entire set of structures.
The first release of every year, in January, will include all structures. April, July and October updates will only contain the structures released during the previous quarter. New subscribers will receive the January release of the current year and all subsequent updates while supplies last.
The index files in the pub/resource sub-directory continue to include all structures in the current PDB FTP site as of that release.
Experimental data files -- NMR Constraints and X-ray Structure Factors -- are released on the same schedule as the structure files: a complete set in January, and incremental updates for the three subsequent quarters.
NOTE: We are out of stock of Release #103, January 2003. New subscribers will be added to the list for Release #107, January 2004.
Questions should be directed to [email protected]. Ordering information is available at http://www.rcsb.org/pdb/cdrom.html.
11-Nov-2003
Lucene Keyword Search Released on the PDB Web Site
After a period of beta testing, the Lucene keyword search engine has replaced the previously-used LDAP keyword search engine to support text searches on the PDB home page, SearchLite, and the "Text Search" field on SearchFields. Lucene uses an index of the remediated mmCIF files to return much more accurate keyword search results.
Lucene supports wildcard searches, phrases, Boolean queries, and offers a spell checker. Options are offered to narrow the scope of the query, for example, to search for author names or PDB IDs; the default is set to search the entire text of the mmCIF file indices. Additionally, partial word and exact word matches are supported; the default is set to perform an exact word match, unless the partial word match option is selected. The home page keyword search will locate exact word matches to a query.
Examples of supported queries can be found on the SearchLite page at http://www.rcsb.org/pdb/searchlite.html, and additional help can be found at http://www.rcsb.org/pdb/help-searchlite.html.
04-Nov-2003
PDB Focus: Deposition and Release Policies
Guidelines for the deposition of coordinate and experimental data have been set by the IUCr, IUPAC-IUBMB-IUPAB, the NIH, and the journals. These policies are detailed at http://deposit.pdb.org/#release.
Depending upon the hold status selected by the depositior, data release occurs when a depositor gives approval (REL), the hold date has expired (HOLD), or the journal article has been published (HPUB).
As of May 6, 2003 (http://www.rcsb.org/pdb/latest_news.html#hpub), there is a one-year limit on the length of a hold period, including HPUBs. If the citation for a structure is not published within the one-year period, depositors will be given the option to either release or withdraw the deposition.
Detailed deposition and release information is available at http://deposit.pdb.org/#release.
28-Oct-2003
Downloadable PDB_EXTRACT Makes Deposition Easier
The software program PDB_EXTRACT was developed to assist depositors in the automatic preparation of crystallographic depositions. This software tool extracts information for deposition from the output files produced by many applications used for structure determination. Current versions of the following programs are supported: HKL 2000, SCALEPACK, d*TREK, SOLVE, MLPHARE, SHARP/autoSHARP, SHELXD/SHELXE/SHELXD/SHELXL, PHASES, SnB, BnP, DM, Solomon, RESOLVE, CNS, REFMAC, RESTRAIN, TNT, and WARP. PDB_EXTRACT will also be part of the CCP4 Program Suite (version 5).
Files produced by PDB_EXTRACT can be edited on a local Linux workstation using the downloadable version of ADIT, which has been extended to provide access to the large amount of data collected by the PDB_EXTRACT program.
PDB_EXTRACT can be downloaded in source and binary versions for Linux, SGI, SUN, OSF and Mac OSX from http://deposit.pdb.org/software/. Source and Linux binary versions of ADIT are also available.
Questions and comments may be sent to [email protected].
21-Oct-2003
Biological Unit Tutorial Now Available from the PDB
An introduction to biological units in the PDB archive is now accessible at http://www.rcsb.org/pdb/biounit_tutorial.html. This useful guide offers definitions of the terms 'asymmetric unit' and 'biological molecule', indicates where information about the biological unit can be found in PDB and mmCIF coordinate files, and describes how the biological unit files in the PDB have been derived.
The biological unit tutorial is also linked from the View Structure and Download/Display File sections of the Structure Explorer page, as well as under PDB WWW User Guides. For more information, please send inquiries to [email protected].
PDB Newsletter 19 Released
The Fall 2003 issue of the PDB Newsletter is now available in HTML format at http://www.rcsb.org/pdb/newsletter/2003q3/. This issue describes PDB activities during the past quarter in the areas of data deposition and processing; data query, reporting, and access; outreach and education. Highlights from this issue include a "Community Focus" feature on Brian W. Matthews, and an "Education Corner" installment by Paul Craig. Subscriptions for the quarterly printed distribution of the PDB Newsletter may be submitted to [email protected].
14-Oct-2003
PDB Poster Prize -- ECM Winner Announced
Thanks to the students and judges who participated in the PDB Poster Prize competition at the ECM meeting. The prize is designed to recognize student poster presentations involving macromolecular crystallography.
The prize was awarded to Carina Lobley for the poster "Structural Studies of the Enzymes of Pantothenate Synthesis" (Carina M.C. Lobley1, Mairi L. Kilkenny1, Florian Schmitzberger1, Michael E. Webb2, Chris Abell2, Alison G. Smith3,1, Tom L. Blundell1; 1Department of Biochemistry, Cambridge; 2University Chemical Laboratory, Cambridge; 3Department of Plant Sciences, Cambridge).
Special thanks to the judges of all of the student posters at ECM -- G. Davies, C. Kenyon, E.F. Garman, and A. Roodt.
The PDB Poster Prize contest will resume in 2004 - further details will be announced in the PDB web site news.
07-Oct-2003
PDB Focus: Searching for Experimental Data Files
The PDB offers several ways to locate experimental data files for structure entries. The SearchFields interface offers an option to narrow a search to only include entries that have experimental data (X-ray structure factors or NMR restraints) available. This option can be activated by selecting "Experimental Data Availability" from the custom options at the bottom of the SearchFields page. To further narrow the search to only structure factors or only constraint data, select the preferred experimental method from the pull down menu in the "Exp. Technique" field. Information about this and other options available on the SearchFields interface can be found on the SearchFields help page.
Experimental data files can be downloaded from the Structure Explorer page, if the experimental data file is available for that structure. Click on the "Structure Factors" or "NMR Restraints" link on the left side of the page to access the experimental data file.
Experimental data files are also available for downloading from the PDB FTP site. Directories for either X-ray structure factors or NMR restraints can be found in the data/structures/all directory, or subdivided by the second and third character of their PDB ID in the data/structures/divided directory. The data/structures/obsolete directory also offers experimental data files for structures that have been removed from the archive.
30-Sept-2003
PDB Education Listserv
A listserv for educators who use the PDB has been established by Dr. Judith Voet, Professor of Biochemistry at Swarthmore College and member of the PDB Advisory Committee. The purpose of this forum is to provide the PDB with feedback on its present usability by students and educators, and ideas for future directions in support of education. If you would like to participate in the discussion, send an e-mail message to [email protected] with the request to subscribe.
 |
|
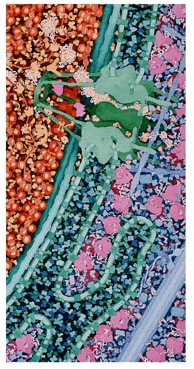
Part of "Macrophage and Bacterium 2,000,000X" by
David S. Goodsell, The Scripps Research Institute |
23-Sep-2003
Illustrations of "Macrophage and Bacterium" by PDB "Molecule of the Month" Author Goodsell Win Award in NSF/Science Visualization Contest
PDB contributor and author of the Molecule of the Month series, Dr. David S. Goodsell, has been awarded second prize in the 2003 Science and Engineering Visualization Challenge for his illustration "Macrophage and Bacterium 2,000,000X". The challenge is a joint project of the National Science Foundation and Science Magazine that encourages and promotes the visual and conceptual beauty of science and engineering.
Dr. Goodsell is an Associate Professor of Molecular Biology at The Scripps Research Institute in La Jolla, California. His research involves computational chemistry and biomolecular computer graphics. Goodsell's artistic talents are multi-faceted, and include many renderings of molecules and cells that are drawn, painted, or generated with the aid of graphics programs. Previously he was awarded the Association of Medical Illustrators Literary Award, among other distinctions he has received. He has contributed many wonderful installments for the PDB Molecule of the Month feature and has authored the popular PDB Molecular Machinery poster--a recent interview with Dr. Goodsell that further describes these efforts can be found in the PDB Newsletter's Spring 2003 issue.
Dr. Goodsell's winning series of three paintings shows a macrophage engulfing a bacterium, including all of the macromolecules in the two cells and in the surrounding blood serum. Goodsell used hundreds of PDB structures for the paintings to get the sizes and shapes of the molecules right: "The Molecular Machinery poster is a good example of the type of information that I start with when I approach a new painting--lots of structures all drawn at a consistent size."
The full winning entry can be viewed at http://www.scripps.edu/pub/goodsell/gallery/macrophagebacterium.html. The original paintings are currently on display in the Center for Integrative Molecular Biosciences at The Scripps Research Institute in La Jolla.
 |
|
This 3-D model of the digestive enzyme chymotrypsin
is part of the "Art of Science" exhibit at Cal State Fullerton. |
16-Sep-2003
PDB Art Part of Molecular Gallery Show at Cal State Fullerton
The PDB's traveling art exhibit is part of the "Art of Science" show at California State University, Fullerton.
Featured are images from the PDB and 3-D models by David Goodsell and Arthur Olson (The Scripps Research Institute). Also included are objects from the Cal State Fullerton Keck Center for Molecular Structure, such as an older generation X-ray camera.
The Art of Science will run through December 19 at the Atrium Gallery in the Paulina June & George Pollak Library. A reception will be held on October 14 at 4pm.
The PDB's Art of Science exhibit includes large-scale depictions of proteins and images and text from the Molecule of the Month series. The PDB would like to see the "Art of Science" travel to other places. If you would be interested in sponsoring this exhibit at your institution, please let us know at [email protected].
09-Sep-2003
PDB Focus: Using rsync to Mirror the PDB FTP Site
One freely available method for establishing and maintaining a local copy of the PDB FTP Site is rsync. The RCSB-created script, rsyncPDB.sh, is a template for using rsync to mirror the FTP archive from an anonymous rsync server. This script can be found at ftp://ftp.rcsb.org/pub/pdb/software/, and the comments in the script explain its usage. Prior to running it, users will need to set three variables in rsyncPDB.sh to suit their local setup. This script is used by the PDB to maintain its FTP mirrors.
General rsync documentation can be found at http://www.samba.org/rsync/. An overview of PDB FTP mirroring procedures is offered at http://www.rcsb.org/pdb/ftpproc.final.html, and the layout of the PDB FTP archive is accessible at http://www.rcsb.org/pdb/ftp_plan.html.
For more information about rsync or mirroring the PDB FTP Site, please send e-mail to [email protected].
02-Sep-2003
Major Enhancements for PDB Web Sites and FTP Archives: Remediated mmCIF Files and Biological Unit Coordinates
Several major enhancements have been implemented, as previously announced -- biological unit coordinates are now accessible in a new directory on the PDB FTP site, and the remediated mmCIF files are now available from the primary PDB web site and its mirrors, and on the PDB FTP site:
mmCIF files
The primary PDB web site and its mirrors now offer the remediated mmCIF files, previously referred to as the "beta" mmCIF files. These files have replaced the set of automatically translated mmCIF files that were created with pdb2cif.pl. The remediated files can be accessed from the Download/Display File section of the Structure Explorer page for any entry, or for a set of query results.
The remediated mmCIF files are also available from the PDB FTP site. The translated files have been removed from the FTP site, and will be made available upon individual request.
For every experimentally-solved structure, both current and obsolete, there is now a remediated mmCIF file in the corresponding directory of the FTP archive in Unix compressed (.Z) format. mmCIF files are not provided for theoretical models. However, the translation software (pdb2cif.pl) is provided at http://www.bernstein-plus-sons.com/software/pdb2cif/ for users who wish to generate the translated mmCIF files.
Biological unit coordinates
Biological unit coordinate files are now available from the FTP archive in the new directory at ftp://ftp.rcsb.org/pub/pdb/data/biounit. These files are accessible in gzipped (.gz) format.
Questions about these new features may be sent to [email protected].
26-Aug-2003
New Version of OpenMMS Toolkit Released
Version 1.5.1 of the OpenMMS software toolkit is now available on the OpenMMS website at http://openmms.sdsc.edu. In addition to the fast database loader, this version contains all of the pdbx fields from the PDB Exchange Dictionary (http://deposit.pdb.org/mmcif/). This includes attributes such as the model number in the atom_site record which is needed for NMR structures. Also available in this release is the "xconv" program which converts mmCIF files to XML files with the standard XML/PDB format.
Questions or comments about the OpenMMS Toolkit may be sent to [email protected].
PDB Poster Prize -- AsCA Winner Announced, Details for ECM Award
Thanks to the students and judges who participated in the PDB Poster Prize competition at the AsCA meeting. The prize is designed to recognize student poster presentations involving macromolecular crystallography.
The prize was awarded to Janet Deane for the poster "Crystal structure of a complex of FLINC4, an intramolecular LMO4:LDB1 complex" (Janet E. Deane, Megan Maher, J. Mitchell Guss, and Jacqueline M. Matthews, School of Molecular and Microbial Biosciences, University of Sydney).
Special thanks to the judges of all of the student posters at AsCA -- Ted Baker (Chair), Peter Colman, Janet S mith, Mark Spackman, Colin Raston, and Yu Wang.
The PDB Poster Prize will also be awarded at the ECM Meeting (August 24-29, Durban, South Africa) meeting.
 |
|
Qualifying posters that display this "PDB" sticker
will be considered for the PDB Poster Prize |
Presenters wishing to enter should collect the distinctive "PDB" sticker from the IUCr exhibit stand and fix it to their poster. The committee appointed to judge the posters will be made up of eminent and willing scientists at the meeting. At a suitable time during the session they will judge the 'best' poster(s) from among those with stickers.
The winner will be notified by e-mail. An announcement will appear on the PDB Web site and in the PDB Newsletter, and in the ACA and IUCr Newsletters.
This year's prize will be signed copies of Biochemistry - Vol. I by Donald and Judith G. Voet and Introduction to Macromolecular Crystallography by Alexander McPherson. The prize will be mailed to the winner after the meeting.
19-Aug-2003
Major Enhancements for Web Sites and FTP Archives Scheduled for Sept. 2
Several major enhancements to the PDB web sites and FTP archives are scheduled to be released with the September 2nd update. Since these changes will involve a large number of files on the FTP archives, we are pre-announcing the planned changes for the benefit of users who mirror the FTP archive:
mmCIF files
The current set of automatically translated mmCIF files will be replaced on all FTP servers and PDB web sites with the new remediated mmCIF files (currently referred to as the "beta" mmCIF files at ftp://beta.rcsb.org/pub/pdb/uniformity/data/):
- All mmCIF files created with pdb2cif.pl will be removed from the FTP archive. They will be archived and made available upon individual request.
- For every experimental structure, both current and obsolete, there will be a new remediated mmCIF file in the corresponding directory of the FTP archive in Unix compressed .Z format.
- No mmCIF files will be provided for theoretical models. However, a link to the translation software (pdb2cif.pl) will be provided for users who wish to generate the translated mmCIF files.
- All PDB web sites will access the new remediated mmCIF files.
Lucene keyword search
The current LDAP keyword search engine that supports text searches on the home page, SearchLite, and the "Text Search" field on SearchFields, will be replaced with the Lucene keyword search engine that is currently in beta testing at http://beta.rcsb.org/pdb/. This will search an index of the remediated mmCIF files and result in much more accurate keyword searches.
Biological unit coordinates
The FTP archive at ftp://ftp.rcsb.org/ will contain the biological unit coordinate files in gzipped .gz format. These files, which are currently served by the beta FTP server, will be kept in a new directory at /pub/pdb/data/biounit.
Questions about these upcoming changes may be sent to [email protected].
 |
|
Ty Gould, PDB Poster Prize winner, and Paul Hubbard, PDB Poster Prize runner up
|
12-Aug-2003
PDB Poster Prize -- ACA Winner Announced, Details for AsCA and ECM Awards
Thanks to the students and judges who participated in the PDB Poster Prize competition at the ACA meeting. The prize is designed to recognize student poster presentations involving macromolecular crystallography.
The first-ever PDB Poster Prize was awarded at the ACA meeting to Ty Gould for the poster "Quorum Sensing Signal Generation by the AHL Synthsase LasI in Pseudomonas aeruginosa Pathogenesis" (T.A. Gould1, R.C. Murphy1, H.P. Schweizer2, M.E.A. Churchill2; 1Dept. of Pharmacology, Univ. of Colorado Health Sciences Center, Denver, CO; 2Dept. of Microbiology, Colorado State Univ., Fort Collins, CO).
A runner-up award was made to Paul Hubbard for the poster "Structure and Catalytic Mechanism of Bacterial 2, 4 - Dienoyl CoA Reductase." (Xiquan Liang3, Horst Schulz3, Jung-Ja Kim, Department of Biochemistry, Medical College of Wisconsin; 3Department of Chemistry, The City University of New York).
Special thanks to the ACA PDB Poster Prize Committee members -- Vivien Yee (Chair), Victor Young, Tom Koetzle, Sylvie Doublie, Marvin L. Hackert, -- and the committee's organizer, Jeanette Krause Bauer.
The PDB Poster Prize will also be awarded at the AsCA (August 10-13 in Broome, Australia) and ECM (August 24-29, Durban, South Africa) meetings.
Presenters wishing to enter should collect the distinctive "PDB" sticker from the IUCr exhibit stand and affix it to their poster. The committee appointed to judge the posters will be made up of eminent and willing scientists at the meeting. At a suitable time during the session they will judge the 'best' poster(s) from among those with stickers.
The winners will be notified by e-mail. An announcement will appear on the PDB web site and in the PDB Newsletter, and in the ACA and IUCr Newsletters.
This year's prize will be signed copies of Biochemistry - Vol. I by Donald and Judith G. Voet and Introduction to Macromolecular Crystallography by Alexander McPherson. The prize will be mailed to the winner after the meetings.
 |
|
Issue 18 shows the new look for the PDB Newsletter
|
05-Aug-2003
PDB Focus: New Features of the PDB Newsletter
Subscribers to the printed version of the PDB Newsletter will have noticed some significant changes in recent issues. The newsletter is now printed in full color and offers attractive molecular images and photos corresponding to the news items in each issue. New columns are included as well: the "PDB Education Corner" describes how educators use the PDB in their curricula, and members of the community and their contributions to the resource are featured each quarter as well.
To receive the printed version of the quarterly PDB Newsletter, please send your request and postal address to [email protected].
29-Jul-2003
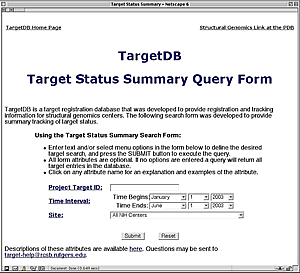
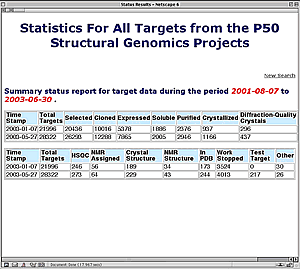
New Summary Report Available from TargetDB
TargetDB (http://targetdb.pdb.org/) is a database of registration and tracking information for structural genomics centers worldwide. TargetDB provides timely status and tracking information on the progress of the production and solution of structures.
The target database can be searched by sequence using FASTA (W.R. Pearson and D.J. Lipman (1988): Improved tools for biological sequence comparison. PNAS 85, pp. 2444-2448). Sequence searches may include the target sequences, PDB sequences, or both. Target sequences may also be searched by contributing site, protein name, project tracking identifier, date of last modification, and the current status of the target (e.g. cloned, expressed, crystallized, ...). Search results may be viewed as HTML reports, FASTA data files, or in XML.
A new feature at the TargetDB site is a search form that can provide summary tracking of target status. This form is now available from TargetDB's main page. Users can search by Target ID, date, or site(s). The summary report includes the number of targets at any given stage in the pipeline. With this new option it is possible to track the progress of a target or of an entire center over a user defined time interval. For instance, reports can be created for each NIH center describing the number of targets in each status category by project year.
Further information about TargetDB and links to structural genomics resources are available at http://www.rcsb.org/pdb/strucgen.html.
|
|
|
|
The new Target Status Summary Query Form (http://targetdb.pdb.org/nih/)
|
Summary report for all NIH centers (January 1 - June 1, 2003) from TargetDB's new Target Status Summary Query Form
|
Scheduled Outage of Select Web Services
As part of ongoing hardware upgrades at the Protein Data Bank's primary web site in San Diego, the following services will be unavailable for several hours on Thursday, July 31:
- the beta web site
- the beta FTP archive (beta mmCIF and XML files)
- biological unit coordinates (from any PDB web site)
- Sting services (from any PDB web site)
We have scheduled a maintenance window of 9 am to 5 pm PDT, Thursday, July 31. However, full functionality may be restored prior to the 5 pm deadline. We apolologize for the inconvenience, and appreciate your patience and understanding.
22-Jul-2003
Demonstrations, Posters, and More: PDB at the ACA Annual Meeting and the 17th Symposium of the Protein Society
The PDB would like to thank those ISMB 2003 attendees who provided valuable feedback at our demonstration session and exhibit during this worthwhile event. PDB staff members will also participate in several other meetings in the near future, including the annual meetings of the Protein Society and the American Crystallographic Association:
Protein Society
The PDB will be presenting a poster "New Features of the Protein Data Bank" at the 17th Symposium of the Protein Society (July 27-29, 2003; Boston, MA). Please stop by and say hello.
ACA Annual Meeting
The PDB will be at the American Crystallographic Association's Annual Meeting at the Northern Kentucky Convention Center in Covington, Kentucky (July 26-31, 2003).
In exhibit booth 112, the PDB will be demonstrating software that is available for use on your own desktop computer. Versions of ADIT, the PDB Validation Suite, PDB_Extract, and other programs can be downloaded from http://deposit.pdb.org/software/.
Also at the meeting, a poster will be presented on "TargetDB: A Target Registration Database for Structural Genomics" during Poster Session II on Monday (P156), and a presentation on "PDB Data Assembly and Validation Tools" will be made as part of the Computational Methods session on Sunday, July 27th at 4:30 in Room 6-8.
15-Jul-2003
Standalone PDB Software Demonstrations at the ACA Annual Meeting
The PDB will be at the American Crystallographic Association's Annual Meeting at the Northern Kentucky Convention Center in Covington, Kentucky (July 26-31, 2003).
In exhibit booth 112, the PDB will be demonstrating software that is available for use on your own desktop computer. Versions of ADIT, the PDB Validation Suite, PDB_Extract, and other programs can be downloaded from http://deposit.pdb.org/software/.
Also at the meeting, a poster will be presented on "TargetDB: A Target Registration Database for Structural Genomics" during Poster Session II on Monday (P156), and a presentation on "PDB Data Assembly and Validation Tools" will be made as part of the Computational Methods session on Sunday, July 27th at 4:30 in Room 6-8.
PDB Poster Prize at ACA -- Instructions for Entering
The PDB Poster Prize will be awarded to the best student poster presentation at this year's ACA meeting.
Presenters wishing to enter should collect the distinctive "PDB" sticker from the PDB exhibit stand (#112) and fix it to their poster. The committee appointed to judge the posters will be made up of eminent and willing scientists at the meeting. At a suitable time during the session they will judge the 'best' poster(s) from among those with stickers.
The winners will be notified by e-mail. An announcement will appear on the PDB Web site and in the PDB Newsletter, and in the ACA and IUCr Newsletters.
This year's prize will be signed copies of Biochemistry - Vol. I by Donald and Judith G. Voet and Introduction to Macromolecular Crystallography by Alexander McPherson. The prize will be mailed to the winner after the ACA meeting.
The PDB Poster Prize will also be awarded at the AsCA (August 10-13 in Broome, Australia) and ECM (August 24-29, Durban, South Africa) meetings.
08-Jul-2003
PDB Focus: How to Access Coordinate Files for Biological Units
Coordinate files for the biological units for applicable structures are accessible from the View Structure and Download/Display File sections of the Structure Explorer pages on the primary PDB web site and its mirrors. The biological unit coordinate files can also be downloaded from the PDB FTP site at ftp://beta.rcsb.org/pub/pdb/biounit/coordinates/ . Subdirectories here are divided by the second and third character of the PDB ID; for example, the biological unit file for PDB entry 4hhb can be found in the /hh/ subdirectory. These files are in gnu-zipped format, and require uncompression. Links to a variety of free uncompression tools can be found at http://www.rcsb.org/pdb/help-general.html#format_structure_compressed .
01-Jul-2003
PDB Newsletter Issue 18 Now Available
The Summer 2003 issue of the PDB Newsletter has been released in HTML format at http://www.rcsb.org/pdb/newsletter/2003q2/. This issue describes the PDB's developments over the past three months in the areas of data deposition and processing; data query, reporting, and access; outreach and education. Subscriptions for the quarterly printed distribution of the PDB Newsletter may be submitted to [email protected].
24-Jun-2003
Biological Unit Files Released on the PDB Web Site
After a period of beta testing, the biological unit images for applicable structures have been implemented on the Structure Explorer pages of the primary PDB Web Site and its mirrors.
The View Structure section of the Structure Explorer now offers still ribbon images of the assumed biological unit(s) for structures, where relevant, in addition to static images of the asymmetric unit. Links to the coordinate files that are used to generate the biological unit images are also accessible here, as well as from the Download/Display File section of the Structure Explorer.
Comments on this feature may be sent to [email protected].
PDB at ISMB 2003
PDB staff members will participate in the 11th International Conference on Intelligent Systems for Molecular Biology (ISMB) to be held June 29-July 3 in Brisbane, Queensland, Australia. New features for query and reporting will be available for testing at PDB's exhibit, booth #10 in the Brisbane Convention & Exhibition Center's Exhibition Hall 1. On Wednesday, July 2, at 9-11am, a demonstration of the re-engineered PDB will be presented in Mezzanine room 1. We look forward to seeing you there!
17-Jun-2003
Enzyme Names and EC Number Query Available From the Structure Explorer Pages on the PDB Web Site
The Summary Information section of the Structure Explorer page for enzymes in the PDB now displays the enzyme name, for entries with complete EC numbers. This section also supports queries for all other PDB entries with the same EC number by clicking on the number shown for that entry. Feedback on these new features is appreciated and may be sent to [email protected].
10-Jun-2003
ADIT Software Available for Download
A standalone version of ADIT for use on your own desktop computer is available for download from http://deposit.pdb.org/software/.
This version has the same features as the web version of ADIT.
ADIT is an integrated software system for editing and checking PDB structure data entries. The system includes tools to help users prepare and check structure depositions. ADIT is currently available in source form and in binary form for Linux platforms.
If you prefer to use the web version of ADIT to deposit your structures, we urge you to run format prechecks and validation prechecks prior to deposition.
03-Jun-2003
PDB Releases XML Data Files for Beta Test
All of the released PDB entries are now available in XML format from the PDB beta FTP site at ftp://beta.rcsb.org/pub/pdb/uniformity/data/XML/. Comments are welcomed on this data.
The XML data files have been created by software translation of the mmCIF data files (ftp://beta.rcsb.org/pub/pdb/uniformity/data/mmCIF/) created as part of the PDB Data Uniformity Project. The mmCIF data files use the data items defined in the PDB Exchange Dictionary (http://deposit.pdb.org/mmcif/). The XML data files conform to an XSD style XML Schema (http://deposit.pdb.org/mmcif/dictionaries/ascii/pdbx-v0.905.xsd) derived from the PDB Exchange Dictionary. As a result, the element and attribute names in the XML data files directly correspond to the item names defined the PDB Exchange Dictionary.
The delivery of PDB data in XML format is the product of a collaboration between the Protein Data Bank Japan (PDBj), the Macromolecular Structure Database (MSD) group at European Bioinformatics Institute (EBI), and the Research Collaboratory for Structural Bioinformatics (RCSB).
27-May-2003
Submission of Structure Factor Data to the PDB
In a recent message to the PDB list server Gerard Kleywegt and Alwyn Jones have requested crystallographers worldwide to deposit structure factor data for existing and future PDB entries. The PDB strongly supports this request.
PDB tries to make the submission of structure factor data as easy as possible. Data can be submitted in ANY format with an accompanying description of content. Structure factor data for new entries can be uploaded along with coordinate data at the time of deposition. For existing entries depositors can simply mail structure factor data to [email protected].
As emphasized in the following message, the structure factor data are an important component of the PDB archive. We ask for the cooperation of all crystallographers to help strengthen the scientific content of the PDB archive by depositing structure factor data for all of their entries both new and old.
-----------------------------------------------------------------------------
Dear colleague !
At present, structure factor data are available for only about *half* of all crystal structures in the PDB. Unless we all make a serious effort now, we must fear that these data will be lost to science for all eternity. Therefore, we would like to encourage all macromolecular crystallographers to check if any of their PDB entries perhaps do not have the associated structure factors deposited. To help you do this, a simple form to query the RCSB database is available at this URL:
http://fsrv1.bmc.uu.se/eds/eds_sos.html
Simply type (a unique part of) your name and hit the "Check" button to get a list of any and all such entries. If there are any, please try and track down the structure factors (on old disks, tapes, or by asking former students and post-docs, etc.) before they are lost forever. If you find any, please send them to the RCSB ([email protected]). (By the way: the most likely future user of deposited structure factor data are you yourself !!)
As you may know, we have been working on creating an archive of electron density maps for all crystal structures in the PDB for which structure factor data have been deposited - the Uppsala Electron Density Server (EDS; URL: http://fsrv1.bmc.uu.se/eds). At present, in about one percent of cases, we are unable to calculate a map at all, and for another ~15% of cases we are unable to reproduce the published R-value to within five percentage-points. The webpage with the search form mentioned above also contains a request for you to help us improve our ~85% success rate with EDS map calculations.
These two initiatives combined will help to preserve and improve the wealth of macromolecular crystallographic data in the public databanks and to make them available and easily accessible to the entire scientific community (cell and molecular biologists, medicinal chemists, crystallographers, etc. etc.) now and in the future.
Thank you for your time and help in advance !!
--Gerard Kleywegt & Alwyn Jones
P.S.: please direct technical correspondence about EDS to [email protected]
P.P.S.: in this request it has been tacitly assumed that coordinates of all published structures have been deposited already. In cases where this is not so, you are of course also strongly encouraged to dig up the models and deposit them together with the corresponding structure factors.
P.P.P.S.: please help this initiative by bringing it to the attention of colleagues who may not read the electronic crystallographic bulletin boards.
Previous Protein Data Bank CD-ROM Sets Available
The PDB has extra CD-ROM sets that were issued prior to January 2003. These sets are copies of the FTP archive that were made at the time of the pressing, and include coordinate and experimental data. Software is not included.
To request any of these CD-ROM sets, send your address and the number of sets you would like to receive to [email protected] or Protein Data Bank, NIST, Mail Stop 8314, Gaithersburg, MD 20899-8314. They will be distributed as long as supplies last on a first come first served basis. The CD-ROM sets will be distributed in reverse order of their date, starting with the most recent.
20-May-2003
Protein Data Bank CD-ROM Sets - First Update Release
The April 2003 release of the PDB CD-ROM sets, issue 104, is an incremental set of 1,317 experimentally determined structures and 23 models. The structures, on one CD_ROM disk, are shipping now.
Structures re-released for any reason between January and April are included in this update. A list of files that have become obsolete since the last update are included so users can update their set of structures.
July and October issues will only contain the structures released during those quarters. New subscribers will receive the January release and all subsequent updates.
The index files in the pub/resource sub-directory continue to include all structures in the current PDB FTP site as of that release.
Experimental data - NMR constraints and X-ray structure factors - will be handled in the same manner as the structures - a complete set in January, and incremental updates for the three subsequent quarters. New subscribers will receive the January release and all updates.
Questions should be directed to [email protected]. Ordering information is available at http://www.rcsb.org/pdb/cdrom.html.
13-May-2003
 |
|
The Art of Science at Purdue U.
|
PDB Art at Purdue University
Images from the PDB's "Art of Science" exhibit are now on display at Purdue University. The PDB installments, along with molecular images from Purdue's Structural Biology Center, are featured in "Watson's Crick", a commons area in the Department of Biological Sciences where local artists present their work.
The exhibit opened on April 11, 2003, and will run until May 17, 2003, in conjunction with the university's Spring Fest events.
The PDB would like to see the "Art of Science" travel to other places. If you would be interested in sponsoring this exhibit at your institution, please let us know at [email protected].
06-May-2003
Clarification of the PDB Policy for "HOLD FOR PUBLICATION" (HPUB) Entries
To insure that coordinate release for structures with "HOLD FOR PUBLICATION" status is consistent with the current PDB Hold Policy, the PDB will place a one-year limit on the length of this hold period. If the citation for a structure is not published within the one-year period, depositors will be given the option to either release or withdraw the deposition.
The one-year limit on the hold period will be applied to new depositions as well as current depositions with "HOLD FOR PUBLICATION" status. Depositors with structures currently held for more than one year are being notified, and given six weeks to either release or withdraw these entries.
29-Apr-2003
PDB Poster Prize
The PDB is pleased to announce the initiation of the PDB Poster Prize, which will recognize student poster presentations involving macromolecular crystallography. The prize will be awarded to the best posters by undergraduate or graduate students at each of the meetings of the IUCr Regional Associates--the American Crystallographic Association (ACA), the Asian Crystallographic Association (AsCA), and the European Crystallographic Association (ECM)--as well as at the IUCr Congress itself. Each award will consist of two educational books; this year's prize will be signed copies of Biochemistry - Vol I by Donald and Judith G. Voet, and Introduction to Macromolecular Crystallography by Alexander McPherson. Winners will be announced on the PDB web site and in the PDB, ACA, and IUCr newsletters.
Details including how to enter can be found at http://www.rcsb.org/pdb/poster_prize.html.
22-Apr-2003
PDB Focus: DNA Day
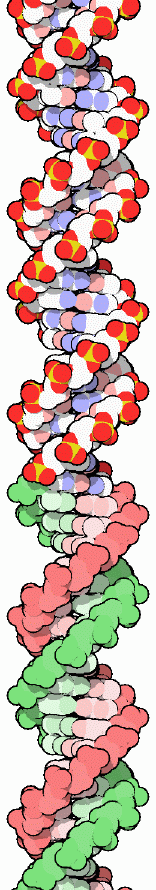 |
||
| Image of B-DNA from the Molecule of the Month installment for Nov., 2001 PDB ID: 1bna H.R. Drew, R.M. Wing, T. Takano, C. Broka, S. Tanaka, K. Itakura, R.E. Dickerson (1981): Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. USA 78, p. 2179. |
||
The National Human Genome Research Institute (http://www.genome.gov/) has a variety of teaching resources for National DNA Day at http://www.genome.gov/10506367.
The Nature Publishing Group has compiled the original articles, historical perspectives, and examinations of DNA in medicine, society, and as a biological molecule in "Double Helix: 50 years of DNA" at http://www.nature.com/nature/dna50/.
The Cold Spring Harbor Laboratory has a Celebration of 50 Years of DNA at http://www.dna50.org/, which provides resources and a schedule of events around the world.
The 50th Anniversary Conference (on April 25th) and other resources from the University of Cambridge are available at http://www.admin.cam.ac.uk/univ/science/dna/.
King's College is sponsoring A Day of Celebrations on April 22 with DNA information at http://www.kcl.ac.uk/depsta/ppro/dna/.
PBS will air a NOVA feature on the "Secret of Photo 51" about Rosalind Franklin's role in the discovery of the structure of DNA.
Other 50th anniversary events, including articles and meetings, are included at http://www.dna50.org.uk/.
An updated and expanded website for the Nucleic Acid Database (NDB), the repository of structural information about nucleic acids, will be released on April 25. The NDB will have a new look and layout, a greatly revised Atlas, a new database that includes X-ray and NMR structures, and a new search engine at http://ndbserver.rutgers.edu/.
The PDB has many education resources related to nucleic acid structure, including DNA's turn as Molecule of the Month in November 2001.
15-Apr-2003
New Features in Beta Testing
PDB users are encouraged to preview the biological unit files, curated (beta) mmCIF files, and redundancy reduction cluster data that are now in a beta testing phase. Comments on these new features are highly appreciated and may be sent to [email protected]:
Biological Unit - Images and Coordinate Files
The biological unit images and corresponding coordinate files for applicable structures are accessible from the Structure Explorer pages on the PDB Beta Web Site at http://beta.rcsb.org/pdb/.
The View Structure section of the Structure Explorer offers still ribbon images of the assumed biological unit(s) for structures, where relevant, in addition to static images of the asymmetric unit. Links to the coordinate files that are used to generate the biological unit images are also accessible here, as well as from the Download/Display File section of the Structure Explorer.
Curated (Beta) mmCIF FilesThe Download/Display File section of the Structure Explorer pages on the Beta Web Site provides links to view or download the curated mmCIF files. These files include remediated data from the Data Uniformity Project. The files follow the latest version of the mmCIF dictionary supplemented by an exchange dictionary developed by the RCSB and the MSD-EBI. This exchange dictionary can be obtained from http://deposit.pdb.org/mmcif/.
The curated mmCIF files for a set of query results can be downloaded by selecting the Download Structures or Sequences option from the pull down menu at the top of the Query Result Browser page.
Curated mmCIF files for all PDB structures are available in gzip (.gz) format at ftp://beta.rcsb.org/pub/pdb/uniformity/data/mmCIF.gz/. UNIX-compressed versions of these files (.Z) are available at ftp://beta.rcsb.org/pub/pdb/uniformity/data/mmCIF/.
New Keyword SearchA much improved keyword search is now available on the beta web site's home page, SearchLite, and the "Text Search" box on SearchFields. This new search engine (powered by Lucene) queries an index derived from the curated mmCIF files, and should return more accurate search results.
08-Apr-2003
PDB Newsletter Issue 17 Released
The Spring 2003 issue of the PDB Newsletter is now available in HTML format. This issue describes the PDB's developments over the past three months in the areas of data deposition and processing; data query, reporting, and access; outreach and education. Subscriptions for the quarterly printed distribution of the PDB Newsletter may be requested by sending your postal address to [email protected].
01-Apr-2003
PDB Focus: David Goodsell and the Molecule of the Month
The Molecule of the Month series explores the functions and significance of selected biological macromolecules for a general audience. These features, written and illustrated by Dr. David S. Goodsell of The Scripps Research Institute, are available here.
Recently, the PDB interviewed Dr. Goodsell to find out how he creates these beautiful and informative works of art and science.
PDB: How did this idea emerge initially?
Goodsell: When I started, I wanted to create a friendly doorway to the PDB. The PDB contains many interesting structures, but
it can be daunting to people who aren't experienced with atomic coordinates and molecular viewers.
One great challenge is the sheer magnitude of the PDB. For instance, if you are interested in hemoglobin,
you are faced with dozens of structures, and it may be difficult to choose one for further exploration.
My goal these days is to present a general introduction to each molecule, and then give a few suggestions for PDB
entries that show the major features of the molecule. A place for visitors to start in their own exploration of these
fascinating molecular machines.
PDB: How do you create the illustrations?
Goodsell: Most of the pictures are created with a computer program that I developed back when I was doing postdoctoral
work with Dr. Art Olson here at The Scripps Research Institute. I've been using this style of illustration--with flat colors
and black outlines--for about 10 years now. I like the way that this style simplifies the molecule, giving a feeling for the
overall shape and form of the molecule, but at the same time you can still see all the individual atoms. On the last page of
each Molecule of the Month--"Exploring the Structure"--I always use RasMol, to give visitors an idea of the kinds of pictures
that they can create themselves with off-the-shelf software.
Proteins are challenging subjects to illustrate. I try to find views that show off the unusual features of the molecules. I like to work with molecules where there is a clear relationship between the structure and the function, such as the way that the ribosome clamps around the messenger RNA or the power stroke motion of myosin. I am also fascinated by the beautiful symmetry of proteins, and always create pictures that highlight this symmetry. Every Molecule of the Month is a new adventure.
PDB: How do you select the featured structures?
Goodsell: I try to pick molecules that play a familiar role in human life and health. My favorites are molecules where we can see
how the molecular structure and function are directly related to something that we experience in our lives. Myosin is a good example--
we can easily imagine those countless little engines crawling up actin as we bend our arm. For each new Molecule of the Month, I try
to pick 4-5 PDB entries that, in my opinion, best show the functional features that I am describing.
PDB: Has it been popular?
Goodsell: Well, I hope so! I have gotten a bunch of great letters from visitors--students, teachers, researchers, and all sorts of
other people. I always like it when people use my pictures in their own assignments or presentations, to aid in their own exploration of the subject.
PDB: What do you plan for the future?
Goodsell: Lots more molecules! I'm planning a new column on hemoglobin with Dr. Shuchismita Dutta (Rutgers-PDB), who helped out on the one on
potassium channels a few months ago--look for it later this Spring. I don't have any plans to enlarge the Molecule of the Month--the PDB is growing
too fast to think of doing anything more comprehensive. I'm planning to keep it small and informal--a new tidbit each month.
25-Mar-2003
Lucene-based Keyword Search on the PDB Beta Web Site
Keyword searches using Lucene can now be performed from the home page, SearchLite, and SearchFields interfaces of the PDB Beta Web Site.
Lucene searches indices of the remediated mmCIF files from the Data Uniformity Project. The use of improved data as a basis for queries, and the use of a ranking system based on finding the keyword in relevant mmCIF categories, facilitate more accurate results than the current keyword search system (LDAP).
The following features are supported by Lucene:
- Wildcard searches: wildcards can be embedded in the center or at the end of a word; e.g., "h*moglobin" will return entries that include "hemoglobin" or "haemoglobin" in their mmCIF files
- Boolean searches: 'and', 'or', 'not' can be used to modify the query; e.g., "calcium and kinase" will return entries that include both "calcium" and "kinase" somewhere in their mmCIF files
- Phrase searches: queries for phrases are supported; e.g., a query for "protein kinase" will return entries that include the phrase "protein kinase" in their mmCIF files
- Queries using parenthesis: queries can be built using parentheses; e.g., "kinase and (calcium or calmodulin)" will return all entries that include kinase, and either calcium or calmodulin, in their mmCIF files
- Spell checker: suggested corrections for misspelled words are presented
The SearchLite and SearchFields pages also offer options to narrow the scope of the query to search specifically for author names or PDB IDs; the default is set to search the entire text of the mmCIF file indices. Additionally, SearchLite and SearchFields support partial word and exact word matches; the default is set to perform an exact word match, unless the partial word match option is selected.
More examples of supported queries can be found on the Beta Web Site's SearchLite page, and corresponding help page.
Comments on this new feature are appreciated, and may be sent to [email protected].
Generator Update at Rutgers-PDB site on March 29, 2003
On Saturday, March 29, 2003, a back up power generator will be installed at Rutgers University. The Rutgers-PDB site will be unavailable from approximately 5:00 AM through 1:00 PM EDT on this date. During this time period, structures can continue to be deposited via ADIT at http://pdbdep.protein.osaka-u.ac.jp/adit/ (Osaka University, Japan) and AutoDep http://autodep.ebi.ac.uk/ (European Bioinformatics Institute, UK).
18-Mar-2003
Redundancy Reduction Cluster Data Now Available for Beta Testing
The results of the weekly clustering of protein chains in the PDB are now available for beta testing at ftp://ftp.rcsb.org/pub/pdb/derived_data/NR/. These clusters are used in the "remove sequence homologs" feature on the PDB web sites. Files that list the clusters and their rankings at 50%, 70% and 90% sequence identity are available. Smaller rank numbers indicate higher (better) ranking. Chains with rank number 1 are ranked as the best representative of their cluster.
The contents of these files and the details of the clustering and ranking are further described at ftp://ftp.rcsb.org/pub/pdb/derived_data/NR/README and in the help documentation.
PDB Focus: Weekly Updates to the PDB
The PDB is updated with new structures each week by Wednesday, 1:00 a.m. Pacific time. This schedule is maintained each week; any changes that may occur are announced in the PDB news. Users can access structures from any previous update using the getPdbUpdate.pl script, which can be found at ftp://ftp.rcsb.org/pub/pdb/software/. Usage of this script is explained at ftp://ftp.rcsb.org/pub/pdb/software/getPdbUpdate.html. An announcement about the latest update is sent every Wednesday morning to the PDB-L discussion forum. To subscribe to the discussion forum, please click here.
11-Mar-2003
Structural Bioinformatics Book Includes Chapters on the PDB
The recently published book, Structural Bioinformatics, includes several chapters about the PDB, which describe its history, function, development, and future goals, as well as the different data formats and protocols used to represent PDB structures:
The PDB Team (2003). The Protein Data Bank. Structural Bioinformatics. P. E. Bourne and H. Weissig. Hoboken, NJ, John Wiley & Sons, Inc. pp. 181-198.
Westbrook, J and Fitzgerald, PM (2003). The PDB format, mmCIF formats and other data formats. Structural Bioinformatics. P. E. Bourne and H. Weissig. Hoboken, NJ, John Wiley & Sons, Inc. pp. 161-179.
Structural Bioinformatics facilitates an understanding of the theories, algorithms, resources, and tools that are used to study biomacromolecular structures such as those included in the PDB. The topics covered -- including proteins, DNA, RNA, carbohydrates, and complex structures -- offer the reader a better understanding of biological function.
Structural Bioinformatics. P. E. Bourne and H. Weissig. Hoboken, NJ, John Wiley & Sons, Inc. (2003)
| Some examples of biological unit images: | |
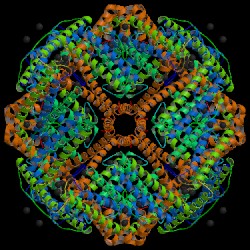 |
|
| PDB ID: 1aew Hempstead, P. D., Yewdall, S. J., Fernie, A. R., Lawson, D. M., Artymiuk, P. J., Rice, D. W., Ford, G. C., Harrison, P. M. (1997): Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution. J. Mol. Biol. 268, p. 424. |
|
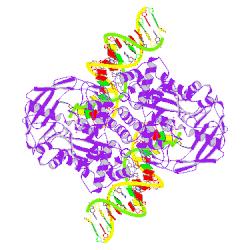 |
|
| PDB ID: 1mm8 Steiniger-White, M., Bhasin, A., Lovell, S., Rayment, I., Reznikoff, W. S. (2002): Evidence for "Unseen" Transposase--DNA Contacts. J. Mol. Biol. 322, p. 971. |
04-Mar-2003
Biological Unit and Curated (Beta) mmCIF Files Now Available from the PDB Beta Web Site
The biological unit images for applicable structures, and curated mmCIF files for all structures, are now accessible from the Structure Explorer pages on the PDB Beta Web Site.
The View Structure section of the Structure Explorer now offers still ribbon images of the assumed biological unit(s) for structures, where relevant, in addition to static images of the asymmetric unit. The interactive molecular viewers available from this page continue to visualize the asymmetric unit. However, links to the coordinate files that are used to generate the biological unit images are also accessible here, as well as from the Download/Display File section of the Structure Explorer.
Additionally, the Download/Display File section now provides links to view or download the curated mmCIF files. These files include remediated data from the Data Uniformity Project. The files follow the latest version of the mmCIF dictionary supplemented by an exchange dictionary developed by the PDB and the Macromolecular Structure Database (MSD) group at the European Bioinformatics Institute. This exchange dictionary can be obtained from http://deposit.pdb.org/mmcif/.
The curated mmCIF files for a set of query results can be downloaded by selecting the Download Structures or Sequences option from the pull down menu at the top of the Query Result Browser page.
Curated mmCIF files for all PDB structures are available in gzip (.gz) format at ftp://beta.rcsb.org/pub/pdb/uniformity/data/mmCIF.gz/. Unix-compressed versions of these files (.Z) remain available at ftp://beta.rcsb.org/pub/pdb/uniformity/data/mmCIF/.
Comments on these new features are appreciated and may be sent to [email protected].
25-Feb-2003
New Distribution Procedure for Protein Data Bank CD-ROM Sets - Starting with Current Issue, 103
The January 2003 release of the PDB CD-ROM sets, issue 103, is a full release of 19,623 experimentally determined structures that were available as of January 1, 2003. The structures, on five CD-ROM disks, are shipping now.
Starting with the April 2003 release, an incremental set of structures released since the January 2003 Issue will be sent to subscribers. Structures re-released for any reason between January and April will be included in this update. A list of files that have become obsolete since the last update will be included so users can update their set of structures.
July and October Issues will only contain the structures released during those quarters. New subscribers will receive the January release and all subsequent updates.
The index files in the pub/resource sub-directory will continue to include all structures in the current PDB FTP site as of that release.
Experimental data - NMR constraints and X-ray structure factors - will be handled in the same manner as the structures - a complete set in January, and incremental updates for the three subsequent quarters.
Questions should be directed to [email protected]. Ordering information is available here.
BioMagResBank Links Included on the Structure Explorer Pages
Links to the BioMagResBank (BMRB) are now included on the Structure Explorer pages for NMR-solved PDB structures that are also available in the BMRB resource. These links return BMRB NMR restraints grid for the particular PDB structure being explored. The BMRB database contains NMR chemical shifts derived from proteins and peptides, reference data, amino acid sequence information, and data describing the source of the protein and the conditions used to study the protein. Images of the structures are also available.
The BMRB links on the Structure Explorer pages are directed to the BMRB site maintained at the University of Wisconsin-Madison, an RCSB partner site.
PDB at the Biophysical Society Meeting
The PDB will participate in the exhibition at the 47th Annual Meeting of the Biophysical Society, to be held March 1-5, in San Antonio, TX. PDB staff will be available at booth #517 to answer questions. We hope to see you there!
18-Feb-2003
PDB Focus: Maintaining a Local PDB FTP Mirror Site
There are several freely available methods for establishing and maintaining a local copy of the PDB FTP Site. The methods described below have the added benefit of preserving the timestamps on files:
- rsyncPDB.sh - This RCSB-created script is a template for using rsync
to mirror the FTP archive from an anonymous rsync server. The script can be found at
ftp://ftp.rcsb.org/pub/pdb/software/,
and the comments in the script explain its usage. Before successfully running it, users will need to
set three variables in rsyncPDB.sh to suit their local setup. This script is now used by the PDB to
maintain its FTP mirrors.
- mirror.pl - This non-RCSB script, which had been used to mirror the PDB FTP archive in the past, is available under the GNU public license from ftp://sunsite.org.uk/packages/mirror. It is recommended to install the ftp.pl_wupatch security patch with the script, also available from this site.
The RCSB-created getPdbUpdate.pl script can also be useful for providers of new FTP mirror sites in obtaining the files from any one particular update. The script can be found at ftp://ftp.rcsb.org/pub/pdb/software/, and its usage is explained at ftp://ftp.rcsb.org/pub/pdb/software/getPdbUpdate.html. Be aware that only LWP::UserAgent, but not wget, preserves the original time stamps of the files.
PDB FTP mirroring procedures are further explained here, and the layout of the PDB FTP archive is accessible here. For more information about mirroring the PDB FTP Site, please send e-mail to [email protected].
11-Feb-2003
Author and Ligand Searches Now Available From the Structure Explorer Pages on the PDB Web Site
The Structure Explorer page for any PDB entry now supports queries for all other entries by a specific primary citation author, or that include a particular ligand listed on that page. These features are available from the Summary Information section of the Structure Explorer page.
By clicking on any individual primary citation author's name, all PDB entries by that author will be returned.
A query for all entries that contain an individual ligand is performed by clicking on any ligand in the "Retrieve all PDB IDs Containing" column in the "HET groups" table.
Feedback on these new features is appreciated and may be sent to [email protected].
04-Feb-2003
PDB Focus: Redundancy Reduction Capability
A subset of structures from which homologous sequences have been largely removed can be obtained from the result list of a query. This option--which is activated by selecting the "remove sequence homologs" option on SearchLite, SearchFields, and the PDB home page-- filters subsets of structures that match a particular query. The default threshold for sequence similarity removal for queries from the home page or SearchLite is 90%; SearchFields provides the option of selecting either 50, 70, or 90% similarity as cut-off values. Users can toggle between the complete set of results and the reduced subset by using the options menu at the top of the Query Result Browser.
Further information about this feature is available in the help documentation.
 |
| Annotators Takashi Kosada (Osaka University) and Bohdan
Schneider (Center for Complex Molecular Systems and Biomolecules in
the Czech Republic) on a visit to the RCSB-Rutgers site. PDB Annotators (top row, left to right): Shri Jain, Anthony Adelakun, Bohdan Schneider; (middle row): Kyle Burkhardt, Shuchismita Dutta, Suzanne Richman; (bottom): Rose Oughtred, Jessica Marvin, Takashi Kosada, Tania Rose Posa |
28-Jan-2003
PDB Focus: ADIT Annotators
PDB data are processed by an international effort. Structures deposited using ADIT are processed by staff from the RCSB (at Rutgers University in New Jersey and remotely at the Center for Complex Molecular Systems and Biomolecules in the Czech Republic) and from the Institute for Protein Research at Osaka University.
Structures are also deposited using AutoDep at the European Bioinformatics Institute (EBI) in the United Kingdom. Data deposited using AutoDep are processed by the EBI.
|
ADIT
|
RCSB Osaka University |
http://pdb.rutgers.edu/adit/ http://pdbdep.protein.osaka-u.ac.jp/adit/ |
|
AutoDep
|
EBI | http://autodep.ebi.ac.uk/ |
21-Jan-2003
PDB Newsletter 16 Now Available
The Winter 2003 issue of the PDB Newsletter has been released in HTML format. This newsletter describes PDB's endeavors and developments over the past three months in the areas of data deposition and processing; data query, reporting, and access; and outreach. This issue will soon be available in PDF and print format as well. Subscriptions for the quarterly printed distribution of the PDB Newsletter may be requested by sending your postal address to [email protected].
14-Jan-2003
PDB Paper Published in Nucleic Acids Research
The PDB recently published a paper, "The Protein Data Bank and structural genomics", in the latest Database Issue of Nucleic Acids Research. This paper describes some of the resources available from the PDB's portal to structural genomics, including the target registration database, TargetDB.
J. Westbrook, Z. Feng, L. Chen, H. Yang, and H.M. Berman (2003): The Protein Data Bank and structural genomics. Nucl. Acids Res. 31, pp. 489-491.
07-Jan-2003
PDB Annotation Manual Online in PDF and PostScript Formats
The manual used as a guide by the PDB ADIT annotators for PDB Data Processing and Annotation is now available in PDF as well as PostScript format.
This document, a reference for the annotation staff, describes how the PDB data processing software system is used to produce the files that are released into the PDB archive. It is available here.














