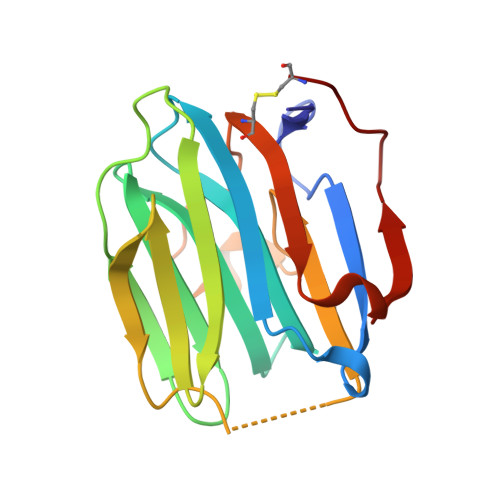
Crystal structure of human sex hormone-binding globulin: steroid transport by a laminin G-like domain.
Grishkovskaya, I., Avvakumov, G.V., Sklenar, G., Dales, D., Hammond, G.L., Muller, Y.A.(2000) EMBO J 19: 504-512
- PubMed: 10675319
- DOI: https://doi.org/10.1093/emboj/19.4.504
- Primary Citation of Related Structures:
1D2S - PubMed Abstract:
Human sex hormone-binding globulin (SHBG) transports sex steroids in blood and regulates their access to target tissues. In biological fluids, SHBG exists as a homodimer and each monomer comprises two laminin G-like domains (G domains). The crystal structure of the N-terminal G domain of SHBG in complex with 5alpha-dihydrotestosterone at 1.55 A resolution reveals both the architecture of the steroid-binding site and the quaternary structure of the dimer. We also show that G domains have jellyroll topology and are structurally related to pentraxin. In each SHBG monomer, the steroid intercalates into a hydrophobic pocket within the beta-sheet sandwich. The steroid and a 20 A distant calcium ion are not located at the dimer interface. Instead, two separate steroid-binding pockets and calcium-binding sites exist per dimer. The structure displays intriguing disorder for loop segment Pro130-Arg135. In all other jellyroll proteins, this loop is well ordered. If modelled accordingly, it covers the steroid-binding site and could thereby regulate access of ligands to the binding pocket.
Organizational Affiliation:
Forschungsgruppe Kristallographie, Max-Delbrück-Center for Molecular Medicine, Robert-Roessle-Strasse 10, D-13092 Berlin, Germany.