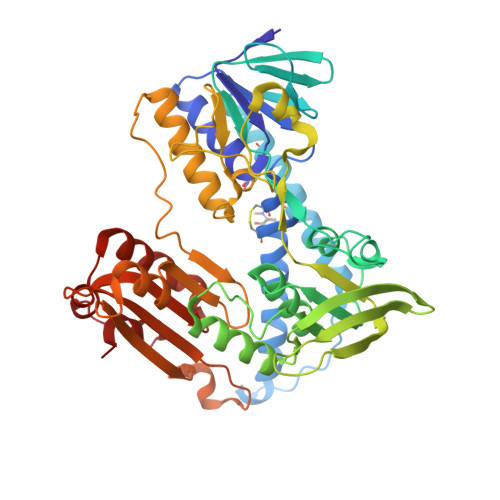
Crystal structure of the antioxidant enzyme glutathione reductase inactivated by peroxynitrite.
Savvides, S.N., Scheiwein, M., Bohme, C.C., Arteel, G.E., Karplus, P.A., Becker, K., Schirmer, R.H.(2002) J Biol Chem 277: 2779-2784
- PubMed: 11705998
- DOI: https://doi.org/10.1074/jbc.M108190200
- Primary Citation of Related Structures:
1K4Q - PubMed Abstract:
As part of our studies on the nitric oxide-related pathology of cerebral malaria, we show that the antioxidative enzyme glutathione reductase (GR) is inactivated by peroxynitrite, with GR from the malarial parasite Plasmodium falciparum being more sensitive than human GR. The crystal structure of modified human GR at 1.9-A resolution provides the first picture of protein inactivation by peroxynitrite and reveals that this is due to the exclusive nitration of 2 Tyr residues (residues 106 and 114) at the glutathione disulfide-binding site. The selective nitration explains the impairment of binding the peptide substrate and thus the nearly 1000-fold decrease in catalytic efficiency (k(cat)/K(m)) of glutathione reductase observed at physiologic pH. By oxidizing the catalytic dithiol to a disulfide, peroxynitrite itself can act as a substrate of unmodified and bisnitrated P. falciparum glutathione reductase.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Oregon State University, Corvallis, Oregon 97331-7305, USA.