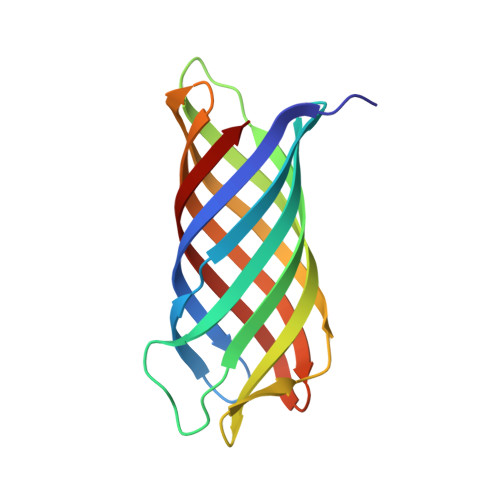
Crystal structure of Neisserial Surface Protein A (NspA), a conserved outer membrane protein with vaccine potential
Vandeputte-Rutten, L., Bos, M.P., Tommassen, J., Gros, P.(2003) J Biol Chem 278: 24825-24830
- PubMed: 12716881
- DOI: https://doi.org/10.1074/jbc.M302803200
- Primary Citation of Related Structures:
1P4T - PubMed Abstract:
The neisserial surface protein A (NspA) from Neisseria meningitidis is a promising vaccine candidate because it is highly conserved among meningococcal strains and induces bactericidal antibodies. NspA is a homolog of the Opa proteins, which mediate adhesion to host cells. Here, we present the crystal structure of NspA, determined to 2.55-A resolution. NspA forms an eight-stranded antiparallel beta-barrel. The four loops at the extracellular side of the NspA molecule form a long cleft, which contains mainly hydrophobic residues and harbors a detergent molecule, suggesting that the protein might function in the binding of hydrophobic ligands, such as lipids. In addition, the structure provides a starting point for structure-based vaccine design.
Organizational Affiliation:
Department of Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Utrecht, The Netherlands.