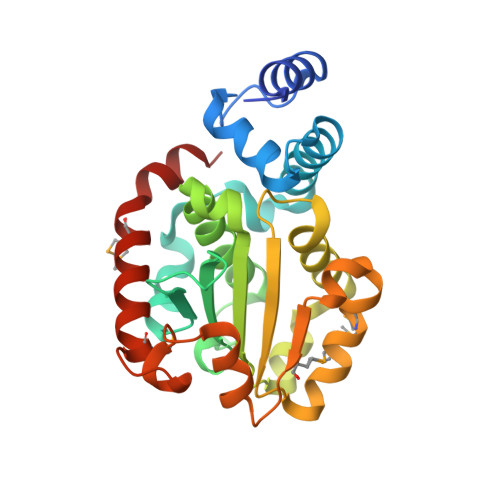
Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency
Min, K.C., Kovall, R.A., Hendrickson, W.A.(2003) Proc Natl Acad Sci U S A 100: 14713-14718
- PubMed: 14657365
- DOI: https://doi.org/10.1073/pnas.2136684100
- Primary Citation of Related Structures:
1R5L - PubMed Abstract:
Human alpha-tocopherol (alpha-T) transfer protein (ATTP) plays a central role in vitamin E homeostasis, preventing degradation of alpha-T by routing this lipophilic molecule for secretion by hepatocytes. Mutations in the gene encoding ATTP have been shown to cause a severe deficiency in alpha-T, which results in a progressive neurodegenerative spinocerebellar ataxia, known as ataxia with vitamin E deficiency (AVED). We have determined the high-resolution crystal structure of human ATTP with (2R,4'R,8'R)-alpha-T in the binding pocket. Surprisingly, the ligand is sequestered deep in the hydrophobic core of the protein, implicating a large structural rearrangement for the entry and release of alpha-T. A comparison to the structure of a related protein, Sec14p, crystallized without a bona fide ligand, shows a possibly relevant open conformation for this family of proteins. Furthermore, of the known mutations that cause AVED, one mutation, L183P, is located directly in the binding pocket. Finally, three mutations associated with AVED involve arginine residues that are grouped together on the surface of ATTP. We propose that this positively charged surface may serve to orient an interacting protein, which might function to regulate the release of alpha-T through an induced change in conformation of ATTP.
Organizational Affiliation:
Howard Hughes Medical Institute and Department of Neurology, Columbia University, New York, NY 10032, USA.