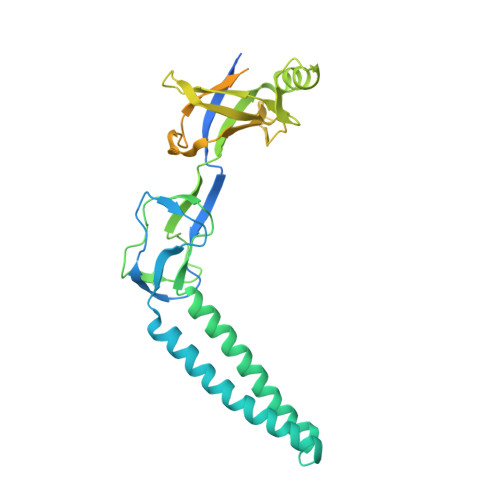
Structure of the periplasmic component of a bacterial drug efflux pump
Higgins, M.K., Bokma, E., Koronakis, E., Hughes, C., Koronakis, V.(2004) Proc Natl Acad Sci U S A 101: 9994-9999
- PubMed: 15226509
- DOI: https://doi.org/10.1073/pnas.0400375101
- Primary Citation of Related Structures:
1T5E - PubMed Abstract:
Multidrug resistance among Gram-negative bacteria is conferred by three-component membrane pumps that expel diverse antibiotics from the cell. These efflux pumps consist of an inner membrane transporter such as the AcrB proton antiporter, an outer membrane exit duct of the TolC family, and a periplasmic protein known as the adaptor. We present the x-ray structure of the MexA adaptor from the human pathogen Pseudomonas aeruginosa. The elongated molecule contains three linearly arranged subdomains; a 47-A-long alpha-helical hairpin, a lipoyl domain, and a six-stranded beta-barrel. In the crystal, hairpins of neighboring MexA monomers pack side-by-side to form twisted arcs. We discuss the implications of the packing of molecules within the crystal. On the basis of the structure and packing, we suggest a model for the key periplasmic interaction between the outer membrane channel and the adaptor protein in the assembled drug efflux pump.
Organizational Affiliation:
Department of Pathology, Cambridge University, Tennis Court Road, Cambridge CB2 1QP, United Kingdom. [email protected]