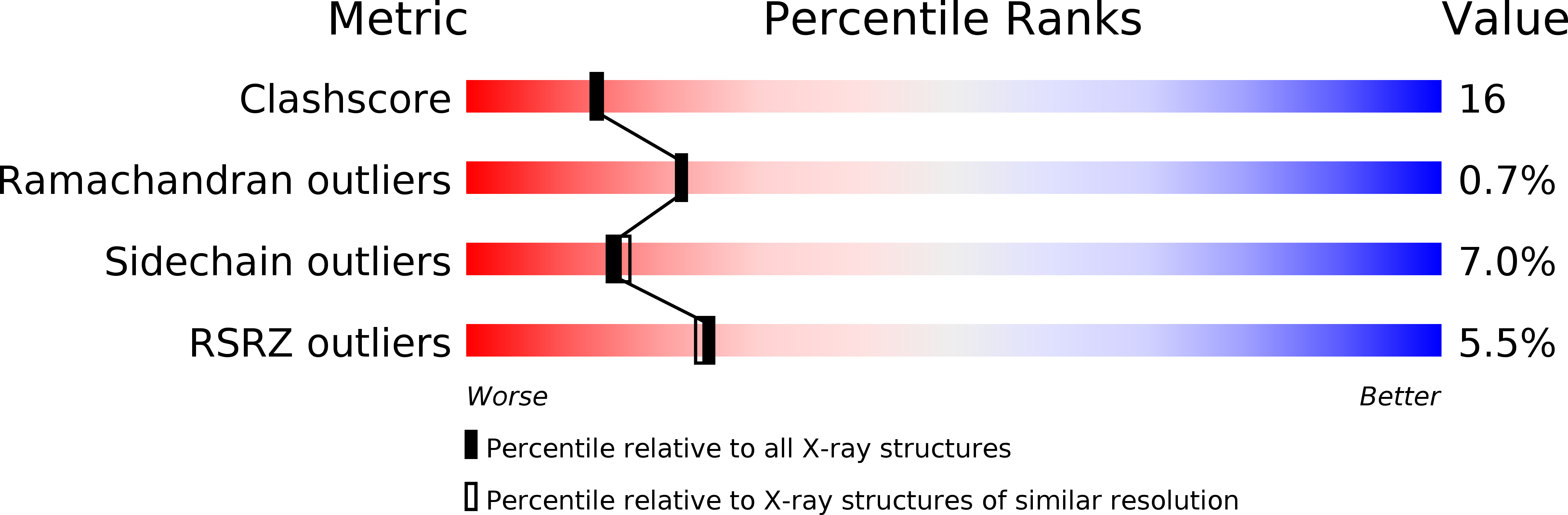
Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity.
Bujacz, G., Alexandratos, J., Wlodawer, A., Merkel, G., Andrake, M., Katz, R.A., Skalka, A.M.(1997) J Biol Chem 272: 18161-18168
- PubMed: 9218451
- DOI: https://doi.org/10.1074/jbc.272.29.18161
- Primary Citation of Related Structures:
1VSH, 1VSI, 1VSJ - PubMed Abstract:
Retroviral integrases (INs) contain two known metal binding domains. The N-terminal domain includes a zinc finger motif and has been shown to bind Zn2+, whereas the central catalytic core domain includes a triad of acidic amino acids that bind Mn2+ or Mg2+, the metal cofactors required for enzymatic activity. The integration reaction occurs in two distinct steps; the first is a specific endonucleolytic cleavage step called "processing," and the second is a polynucleotide transfer or "joining" step. Our previous results showed that the metal preference for in vitro activity of avian sarcoma virus IN is Mn2+ > Mg2+ and that a single cation of either metal is coordinated by two of the three critical active site residues (Asp-64 and Asp-121) in crystals of the isolated catalytic domain. Here, we report that Ca2+, Zn2+, and Cd2+ can also bind in the active site of the catalytic domain. Furthermore, two zinc and cadmium cations are bound at the active site, with all three residues of the active site triad (Asp-64, Asp-121, and Glu-157) contributing to their coordination. These results are consistent with a two-metal mechanism for catalysis by retroviral integrases. We also show that Zn2+ can serve as a cofactor for the endonucleolytic reactions catalyzed by either the full-length protein, a derivative lacking the N-terminal domain, or the isolated catalytic domain of avian sarcoma virus IN. However, polynucleotidyl transferase activities are severely impaired or undetectable in the presence of Zn2+. Thus, although the processing and joining steps of integrase employ a similar mechanism and the same active site triad, they can be clearly distinguished by their metal preferences.
Organizational Affiliation:
Macromolecular Structure Laboratory, NCI-Frederick Cancer Research and Development Center, ABL-Basic Research Program, Frederick, Maryland 21702, USA.