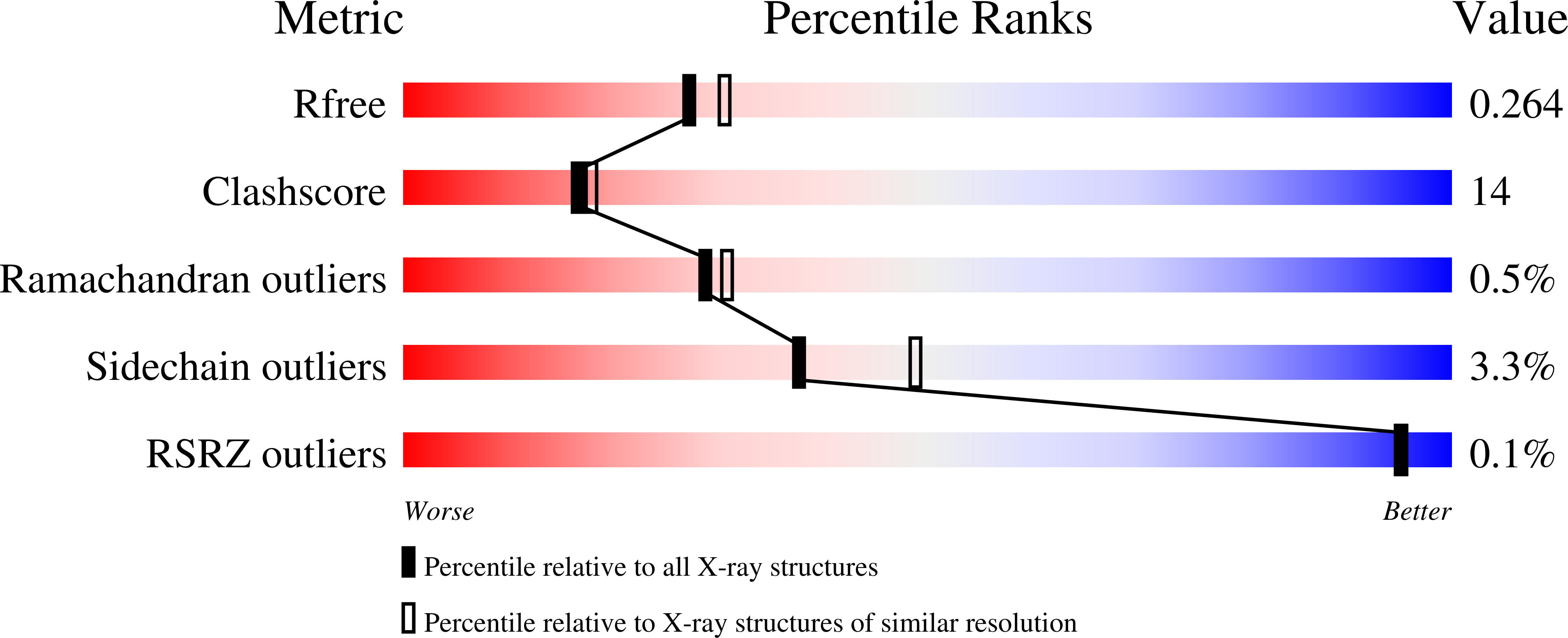
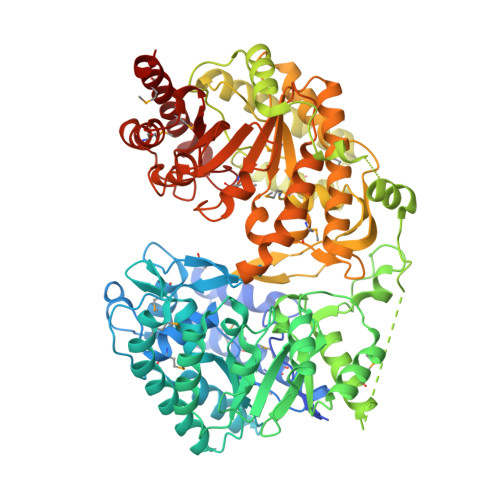
Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication
Pejchal, R., Ludwig, M.L.(2005) PLoS Biol 3: e31
- PubMed: 15630480
- DOI: https://doi.org/10.1371/journal.pbio.0030031
- Primary Citation of Related Structures:
1T7L, 1XDJ, 1XPG, 1XR2 - PubMed Abstract:
Cobalamin-independent methionine synthase (MetE) catalyzes the transfer of a methyl group from methyltetrahydrofolate to L-homocysteine (Hcy) without using an intermediate methyl carrier. Although MetE displays no detectable sequence homology with cobalamin-dependent methionine synthase (MetH), both enzymes require zinc for activation and binding of Hcy. Crystallographic analyses of MetE from T. maritima reveal an unusual dual-barrel structure in which the active site lies between the tops of the two (betaalpha)(8) barrels. The fold of the N-terminal barrel confirms that it has evolved from the C-terminal polypeptide by gene duplication; comparisons of the barrels provide an intriguing example of homologous domain evolution in which binding sites are obliterated. The C-terminal barrel incorporates the zinc ion that binds and activates Hcy. The zinc-binding site in MetE is distinguished from the (Cys)(3)Zn site in the related enzymes, MetH and betaine-homocysteine methyltransferase, by its position in the barrel and by the metal ligands, which are histidine, cysteine, glutamate, and cysteine in the resting form of MetE. Hcy associates at the face of the metal opposite glutamate, which moves away from the zinc in the binary E.Hcy complex. The folate substrate is not intimately associated with the N-terminal barrel; instead, elements from both barrels contribute binding determinants in a binary complex in which the folate substrate is incorrectly oriented for methyl transfer. Atypical locations of the Hcy and folate sites in the C-terminal barrel presumably permit direct interaction of the substrates in a ternary complex. Structures of the binary substrate complexes imply that rearrangement of folate, perhaps accompanied by domain rearrangement, must occur before formation of a ternary complex that is competent for methyl transfer.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, Ann Arbor, USA.