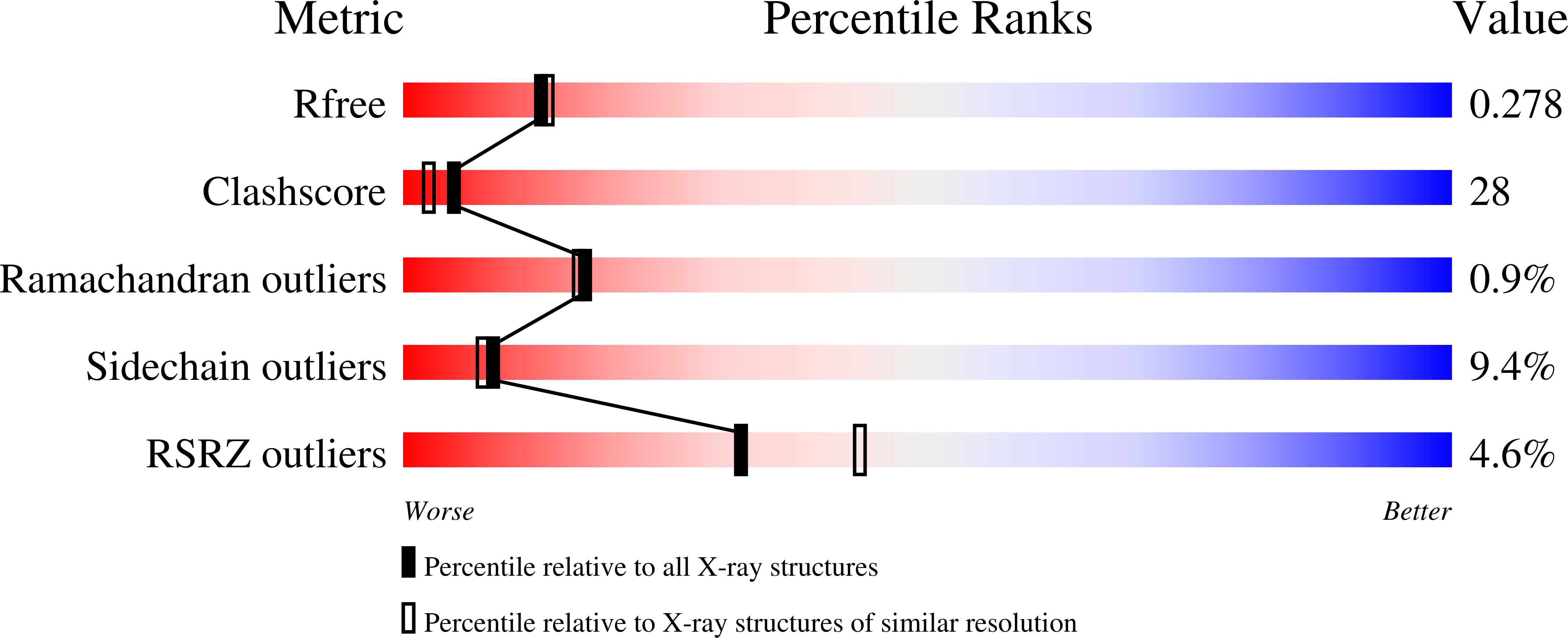
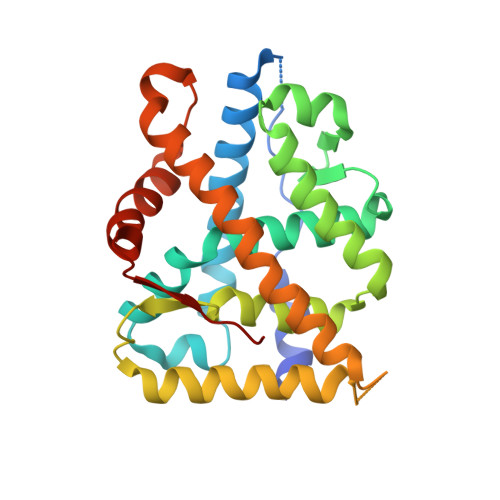
Crystal structure of a mutant mineralocorticoid receptor responsible for hypertension
Fagart, J., Huyet, J., Pinon, G.M., Rochel, M., Mayer, C., Rafestin-Oblin, M.E.(2005) Nat Struct Mol Biol 12: 554-555
- PubMed: 15908963
- DOI: https://doi.org/10.1038/nsmb939
- Primary Citation of Related Structures:
1Y9R, 1YA3 - PubMed Abstract:
The S810L mutation within the human mineralocorticoid receptor (MR S810L) induces severe hypertension and switches progesterone from antagonist to agonist. Here we report the crystal structures of the ligand-binding domain of MR S810L in complex with progesterone and deoxycorticosterone, an agonist of both wild-type and mutant MRs. These structures, the first for MR, identify the specific contacts created by Leu810 and clarify the mechanism of activation of MR S810L.
Organizational Affiliation:
INSERM U478, Faculté de Médecine Xavier Bichat, 16 rue Henri Huchard, B.P. 416, 75870 Paris Cedex 18, France. [email protected]