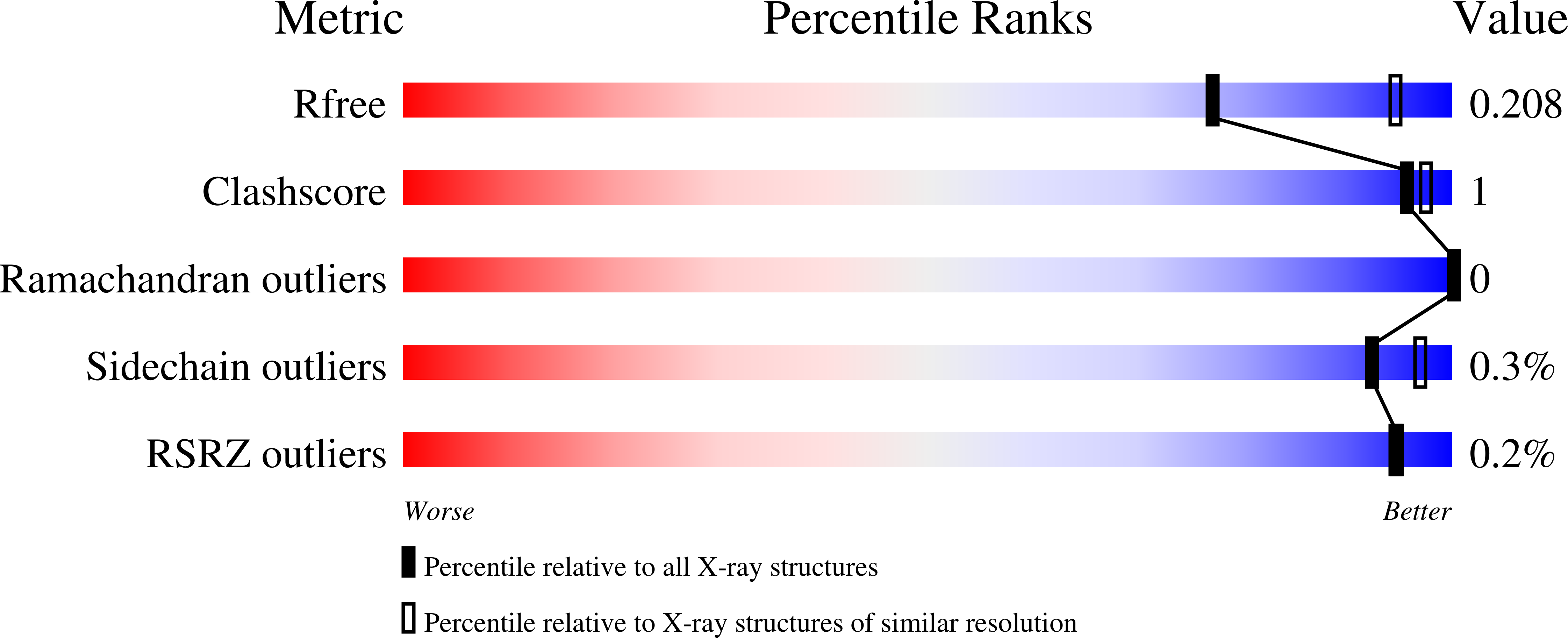
Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Graham, S.C., Bond, C.S., Freeman, H.C., Guss, J.M.(2005) Biochemistry 44: 13820-13836
- PubMed: 16229471
- DOI: https://doi.org/10.1021/bi0512849
- Primary Citation of Related Structures:
1W2M, 1W7V, 1WBQ, 1WL6, 1WL9, 1WLR, 2BH3, 2BHA, 2BHB, 2BHC, 2BHD, 2BN7 - PubMed Abstract:
The effect of metal substitution on the activity and structure of the aminopeptidase P (APPro) from Escherichia coli has been investigated. Measurements of activity in the presence of Mn2+, Mg2+, Zn2+, Na+, and Ca2+ show that significant activity is seen only in the Mn-bound form of the enzyme. The addition of Zn2+ to [MnMn(APPro)] is strongly inhibitory. Crystal structures of [MnMn(APPro)], [MgMg(APPro)], [ZnZn(APPro)], [ZnMg(APPro)], [Ca_(APPro)], [Na_(APPro)], and [apo(APPro)] were determined. The structures of [Ca_(APPro)] and [Na_(APPro)] have a single metal atom at their active site. Surprisingly, when a tripeptide substrate (ValProLeu) was soaked into [Na_(APPro)] crystals in the presence of 200 mM Mg2+, the structure had substrate, but no metal, bound at the active site. The structure of apo APPro complexed with ValProLeu shows that the N-terminal amino group of a substrate can be bound at the active site by carboxylate side chains that normally bind the second metal atom, providing a model for substrate binding in a single-metal active enzyme. Structures of [MnMn(APPro)] and [ZnZn(APPro)] complexes of ProLeu, a product inhibitor, in the presence of excess Zn reveal a third metal-binding site, formed by two conserved His residues and the dipeptide inhibitor. A Zn atom bound at such a site would stabilize product binding and enhance inhibition.
Organizational Affiliation:
School of Molecular and Microbial Biosciences, University of Sydney, NSW 2006, Australia.