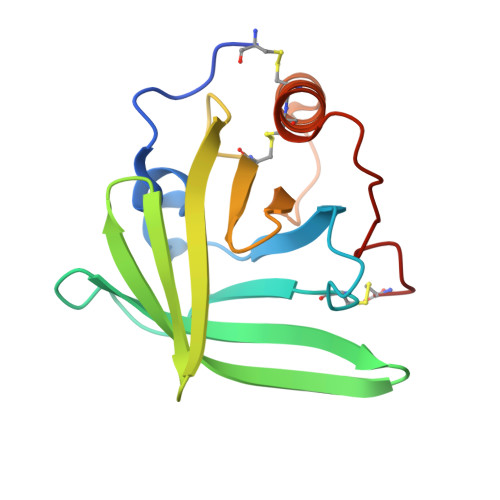
The Structure of Omci, a Novel Lipocalin Inhibitor of the Complement System.
Roversi, P., Lissina, O., Johnson, S., Ahmat, N., Paesen, G.C., Ploss, K., Boland, W., Nunn, M.A., Lea, S.M.(2007) J Mol Biol 369: 784
- PubMed: 17445829
- DOI: https://doi.org/10.1016/j.jmb.2007.03.064
- Primary Citation of Related Structures:
2CM4, 2CM9 - PubMed Abstract:
The complement (C) system is a potent innate immune defence system against parasites. We have recently characterised and expressed OmCI, a 16 kDa protein derived from the soft tick Ornithodoros moubata that specifically binds C5, thereby preventing C activation. The structure of recombinant OmCI determined at 1.9 A resolution confirms a lipocalin fold and reveals that the protein binds a fatty acid derivative that we have identified by mass spectrometry as ricinoleic acid. We propose that OmCI could sequester one of the fatty acid-derived inflammatory modulators from the host plasma, thereby interfering with the host inflammatory response to the tick bite. Mapping of sequence differences between OmCI and other tick lipocalins with different functions, combined with biochemical investigations of OmCI activity, supports the hypothesis that OmCI acts by preventing interaction with the C5 convertase, rather than by blocking the C5a cleavage site.
Organizational Affiliation:
Sir William Dunn School of Pathology, University of Oxford, Oxford OX1 3RE, England, UK.