Enhanced major histocompatibility complex class I binding and immune responses through anchor modification of the non-canonical tumour-associated mucin 1-8 peptide
Lazoura, E., Lodding, J., Farrugia, W., Ramsland, P.A., Stevens, J., Wilson, I.A., Pietersz, G.A., Apostolopoulos, V.(2006) Immunology 119: 306-316
- PubMed: 17067310
- DOI: https://doi.org/10.1111/j.1365-2567.2006.02434.x
- Primary Citation of Related Structures:
2FO4 - PubMed Abstract:
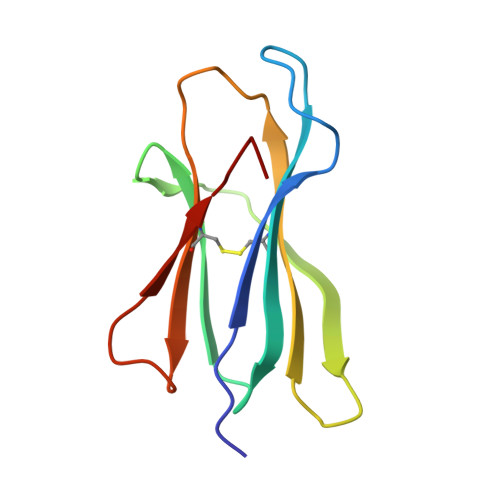
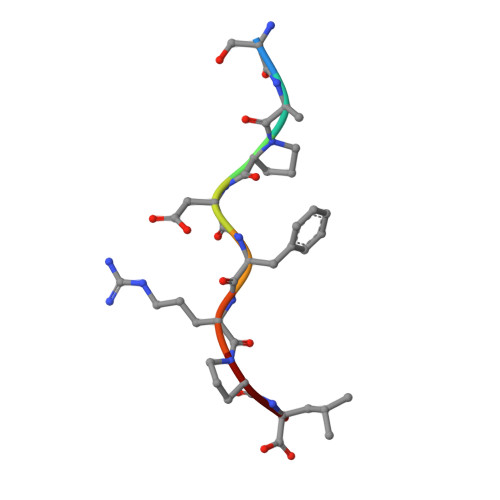
Designing peptide-based vaccines for therapeutic applications in cancer immunotherapy requires detailed knowledge of the interactions between the antigenic peptide and major histocompatibility complex (MHC) in addition to that between the peptide-MHC complex and the T-cell receptor. Past efforts to immunize with high-affinity tumour-associated antigenic peptides have not been very immunogenic, which may be attributed to the lack of T cells to these peptides, having been deleted during thymic development. For this reason, low-to-medium affinity non-canonical peptides represent more suitable candidates. However, in addition to the difficulty in identifying such antigens, peptide binding to MHC, and hence its ability to induce a strong immune response, is limited. Therefore, to enhance binding to MHC and improve immune responses, anchor modifications of non-canonical tumour-associated peptides would be advantageous. In this study, the non-canonical tumour-associated peptide from MUC1, MUC1-8 (SAPDTRPA), was modified at the MHC anchor residues to SAPDFRPL (MUC1-8-5F8L) and showed enhanced binding to H-2Kb and improved immune responses. Furthermore, the crystal structure of MUC1-8-5F8L in complex with H-2Kb was determined and it revealed that binding of the peptide to MHC is similar to that of the canonical peptide OVA8 (SIINFEKL).
Organizational Affiliation:
Burnet Institute at Austin, Immunology and Vaccine Laboratory, Heidelberg, VIC, Australia. [email protected]