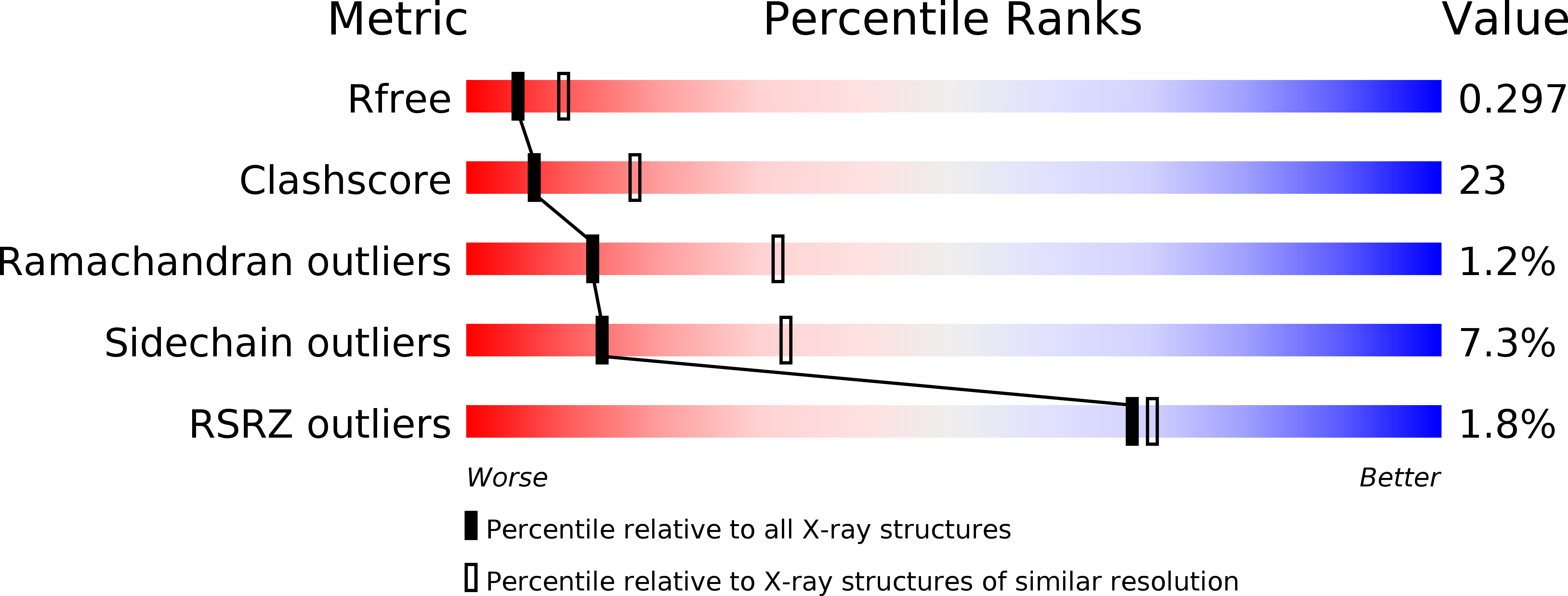
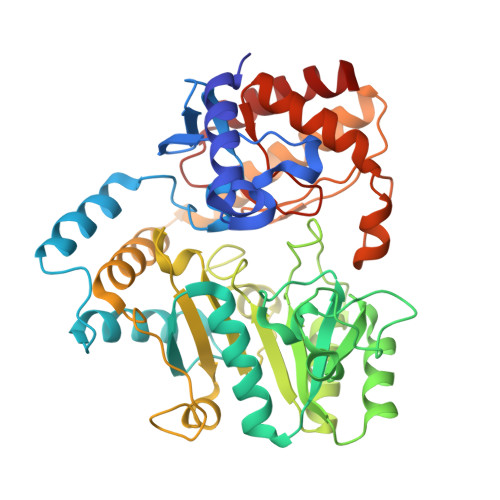
Intersubunit signaling in glutamate-1-semialdehyde-aminomutase.
Stetefeld, J., Jenny, M., Burkhard, P.(2006) Proc Natl Acad Sci U S A 103: 13688-13693
- PubMed: 16954186
- DOI: https://doi.org/10.1073/pnas.0600306103
- Primary Citation of Related Structures:
2HOY, 2HOZ, 2HP1, 2HP2 - PubMed Abstract:
Enzymes are highly dynamic and tightly controlled systems. However, allosteric communication linked to catalytic turnover is poorly understood. We have performed an integrated approach to trap several catalytic intermediates in the alpha2-dimeric key enzyme of chlorophyll biosynthesis, glutamate-1-semialdehyde aminomutase. Our data reveal an active-site "gating loop," which undergoes a dramatic conformational change during catalysis, that is simultaneously open in one subunit and closed in the other. This loop movement requires a beta-sheet-to-alpha-helix transition to assume the closed conformation, thus facilitating transport of substrate toward, and concomitantly forming, an integral part of the active site. The accompanying intersubunit cross-talk, which controls negative cooperativity between the allosteric pair, was explored at the atomic level. The central elements of the communication triad are the cofactor bound to different catalytic intermediates, the interface helix, and the gating loop. Together, they form a molecular switch in which the cofactor acts as a central signal transmitter linking the subunit interface with the gating loop.
Organizational Affiliation:
Department of Structural Biology and M. E. Müller Institute for Structural Biology, Biozentrum Universität Basel, Klingelbergstrasse 70, 4056 Basel, Switzerland. [email protected]