Structural basis of poly(3-hydroxybutyrate) hydrolysis by PhaZ7 depolymerase from Paucimonas lemoignei.
Papageorgiou, A.C., Hermawan, S., Singh, C.B., Jendrossek, D.(2008) J Mol Biol 382: 1184-1194
- PubMed: 18706425
- DOI: https://doi.org/10.1016/j.jmb.2008.07.078
- Primary Citation of Related Structures:
2VTV - PubMed Abstract:
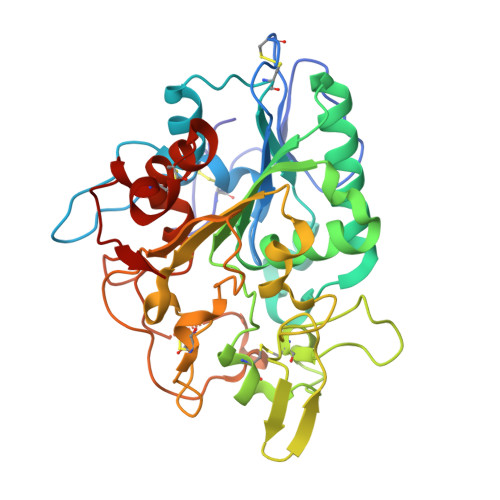
The crystal structure of poly(3-hydroxybutyrate) (PHB) depolymerase PhaZ7 purified from Paucimonas lemoignei was determined at 1.90 A resolution. The structure consists of a single domain with an alpha/beta hydrolase fold in its core. The active site is analogous to that of serine esterases/lipases and is characterized by the presence of a catalytic triad comprising Ser136, Asp242, and His306. Comparison with other structures in the Protein Data Bank showed a high level of similarity with the Bacillus subtilis lipase LipA (RMSD, 1.55 A). Structural comparison with Penicillium funiculosum PHB depolymerase, the only PHB depolymerase whose structure is already known, revealed significant differences, resulting in an RMSD of 2.80-3.58 A. The two enzymes appear to utilize different types of solvent-exposed residues for biopolymer binding, with aliphatic and hydroxyl residues used in P. funiculosum PHB depolymerase and aromatic residues in PhaZ7. Moreover, the active site of P. funiculosum PHB depolymerase is accessible to the substrate in contrast to the active site of PhaZ7, which is buried. Hence, considerable conformational changes are required in PhaZ7 for the creation of a channel leading to the active site. Taken together, the structural data suggest that PhaZ7 and P. funiculosum PHB depolymerase have adopted different strategies for effective substrate binding in response to their diverse substrate specificity and the lack of a substrate-binding domain.
Organizational Affiliation:
Turku Centre for Biotechnology, University of Turku and Abo Akademi University, BioCity, Turku 20521, Finland. [email protected]