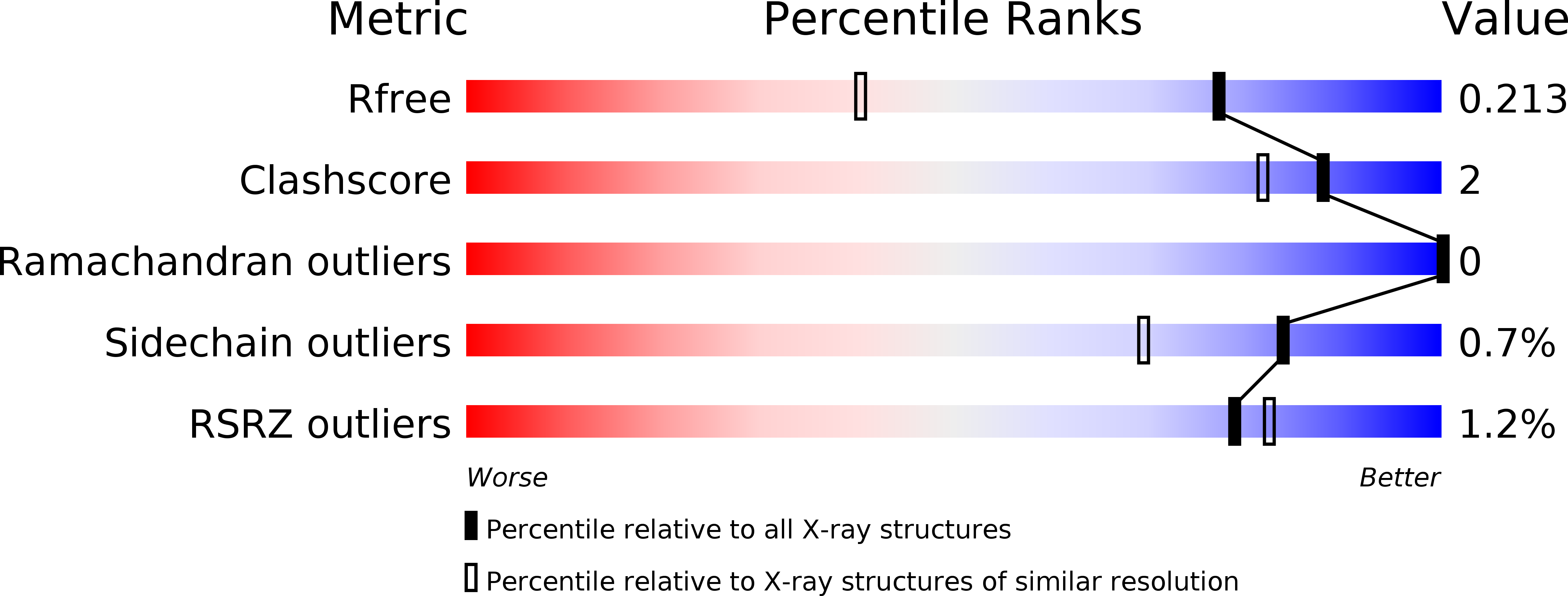
The Aspergillus Fumigatus Sialidase is a Kdnase: Structural and Mechanistic Insights.
Telford, J.C., Yeung, J.H.F., Xu, G., Kiefel, M.J., Watts, A.G., Hader, S., Chan, J., Bennet, A.J., Moore, M.M., Taylor, G.L.(2011) J Biol Chem 286: 10783
- PubMed: 21247893
- DOI: https://doi.org/10.1074/jbc.M110.207043
- Primary Citation of Related Structures:
2XCY, 2XZI, 2XZJ, 2XZK - PubMed Abstract:
Aspergillus fumigatus is a filamentous fungus that can cause severe respiratory disease in immunocompromised individuals. A putative sialidase from A. fumigatus was recently cloned and shown to be relatively poor in cleaving N-acetylneuraminic acid (Neu5Ac) in comparison with bacterial sialidases. Here we present the first crystal structure of a fungal sialidase. When the apo structure was compared with bacterial sialidase structures, the active site of the Aspergillus enzyme suggested that Neu5Ac would be a poor substrate because of a smaller pocket that normally accommodates the acetamido group of Neu5Ac in sialidases. A sialic acid with a hydroxyl in place of an acetamido group is 2-keto-3-deoxynononic acid (KDN). We show that KDN is the preferred substrate for the A. fumigatus sialidase and that A. fumigatus can utilize KDN as a sole carbon source. A 1.45-Å resolution crystal structure of the enzyme in complex with KDN reveals KDN in the active site in a boat conformation and nearby a second binding site occupied by KDN in a chair conformation, suggesting that polyKDN may be a natural substrate. The enzyme is not inhibited by the sialidase transition state analog 2-deoxy-2,3-dehydro-N-acetylneuraminic acid (Neu5Ac2en) but is inhibited by the related 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid that we show bound to the enzyme in a 1.84-Å resolution crystal structure. Using a fluorinated KDN substrate, we present a 1.5-Å resolution structure of a covalently bound catalytic intermediate. The A. fumigatus sialidase is therefore a KDNase with a similar catalytic mechanism to Neu5Ac exosialidases, and this study represents the first structure of a KDNase.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St. Andrews, St. Andrews, Fife KY16 9ST, United Kingdom.