Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
Martins, B.M., Blaser, M., Feliks, M., Ullmann, G.M., Buckel, W., Selmer, T.(2011) J Am Chem Soc 133: 14666
- PubMed: 21823587
- DOI: https://doi.org/10.1021/ja203344x
- Primary Citation of Related Structures:
2Y8N, 2YAJ - PubMed Abstract:
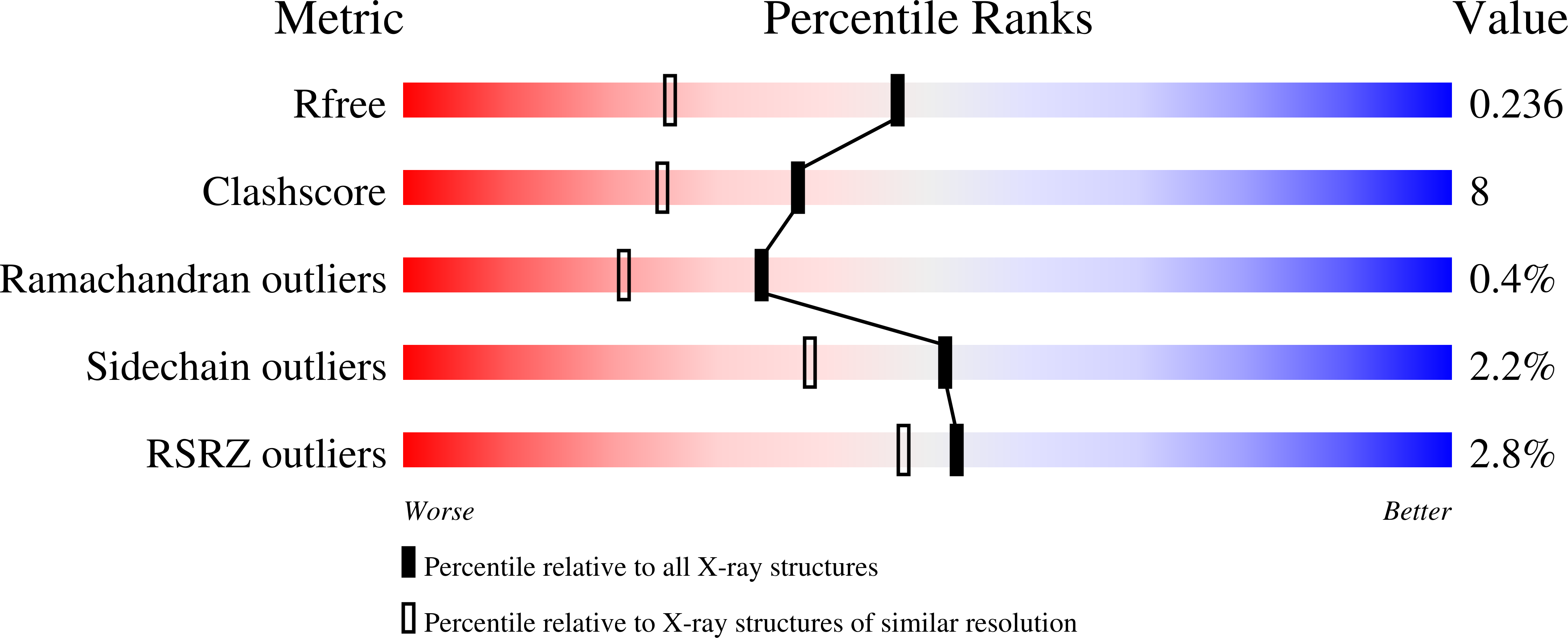
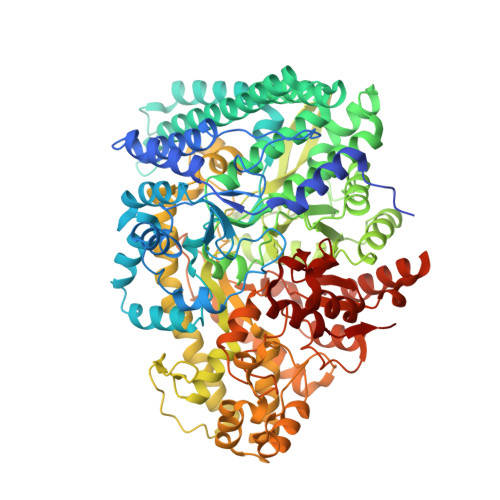
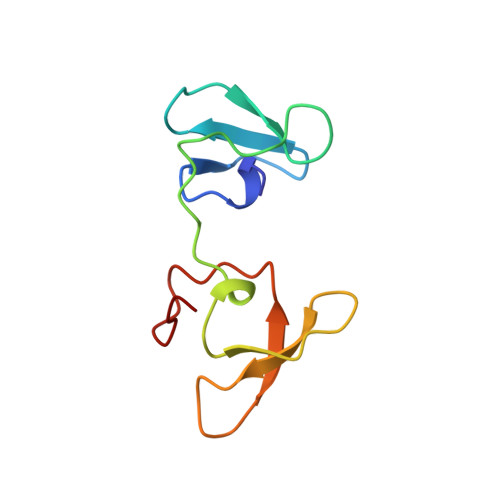
4-Hydroxyphenylacetate decarboxylase is a [4Fe-4S] cluster containing glycyl radical enzyme proposed to use a glycyl/thiyl radical dyad to catalyze the last step of tyrosine fermentation in clostridia. The decarboxylation product p-cresol (4-methylphenol) is a virulence factor of the human pathogen Clostridium difficile . Here we describe the crystal structures at 1.75 and 1.81 Å resolution of substrate-free and substrate-bound 4-hydroxyphenylacetate decarboxylase from the related Clostridium scatologenes . The structures show a (βγ)(4) tetramer of heterodimers composed of a catalytic β-subunit harboring the putative glycyl/thiyl dyad and a distinct small γ-subunit with two [4Fe-4S] clusters at 40 Å distance from the active site. The γ-subunit comprises two domains displaying pseudo-2-fold symmetry that are structurally related to the [4Fe-4S] cluster-binding scaffold of high-potential iron-sulfur proteins. The N-terminal domain coordinates one cluster with one histidine and three cysteines, and the C-terminal domain coordinates the second cluster with four cysteines. Whereas the C-terminal cluster is buried in the βγ heterodimer interface, the N-terminal cluster is not part of the interface. The previously postulated decarboxylation mechanism required the substrate's hydroxyl group in the vicinity of the active cysteine residue. In contrast to expectation, the substrate-bound state shows a direct interaction between the substrate's carboxyl group and the active site Cys503, while His536 and Glu637 at the opposite side of the active site pocket anchor the hydroxyl group. This state captures a possible catalytically competent complex and suggests a Kolbe-type decarboxylation for p-cresol formation.
Organizational Affiliation:
Institute für Biologie, Strukturbiologie/Biochemie, Humboldt-Universität zu Berlin, D-10115 Berlin, Germany. [email protected]