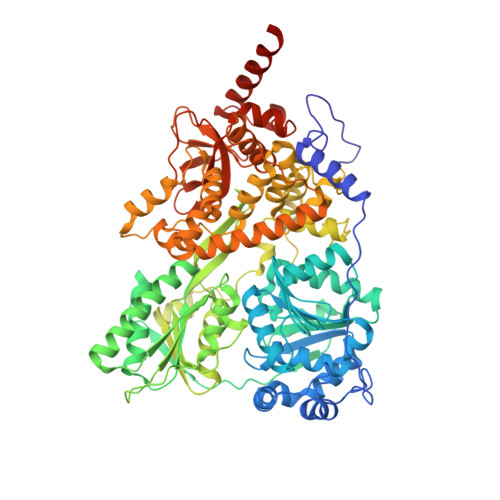
Structural basis for the function of DEAH helicases
He, Y., Andersen, G.R., Nielsen, K.H.(2010) EMBO Rep 11: 180-186
- PubMed: 20168331
- DOI: https://doi.org/10.1038/embor.2010.11
- Primary Citation of Related Structures:
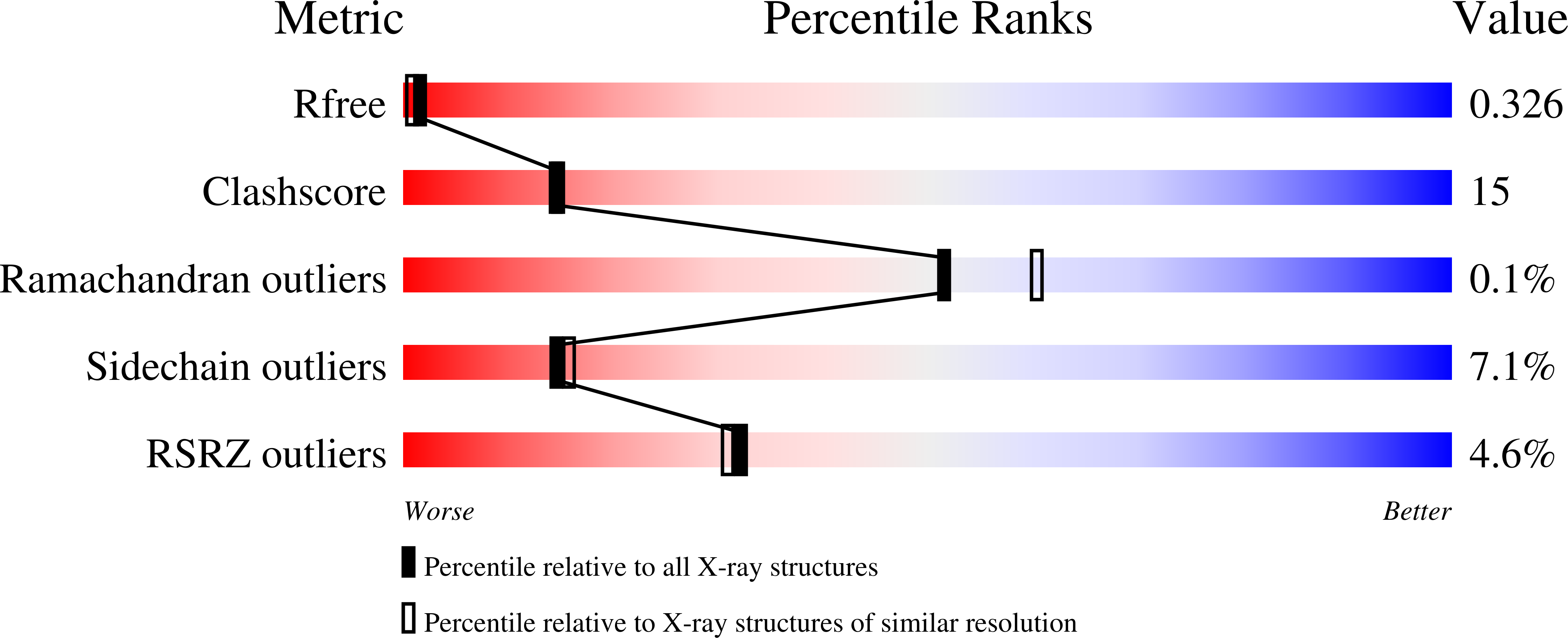
3KX2 - PubMed Abstract:
DEAH helicases participate in pre-messenger RNA splicing and ribosome biogenesis. The structure of yeast Prp43p-ADP reveals the homology of DEAH helicases to DNA helicases and the presence of an oligonucleotide-binding motif. A beta-hairpin from the second RecA domain is wedged between two carboxy-terminal domains and blocks access to the occluded RNA binding site formed by the RecA domains and a C-terminal domain. ATP binding and hydrolysis are likely to induce conformational changes in the hairpin that are important for RNA unwinding or ribonucleoprotein remodelling. The structure of Prp43p provides the framework for functional and genetic analysis of all DEAH helicases.
Organizational Affiliation:
Department of Molecular Biology, Aarhus University, Aarhus, Denmark.