A Missing Enzyme in Thiamin Thiazole Biosynthesis: Identification of TenI as a Thiazole Tautomerase.
Hazra, A.B., Han, Y., Chatterjee, A., Zhang, Y., Lai, R.Y., Ealick, S.E., Begley, T.P.(2011) J Am Chem Soc 133: 9311-9319
- PubMed: 21534620
- DOI: https://doi.org/10.1021/ja1110514
- Primary Citation of Related Structures:
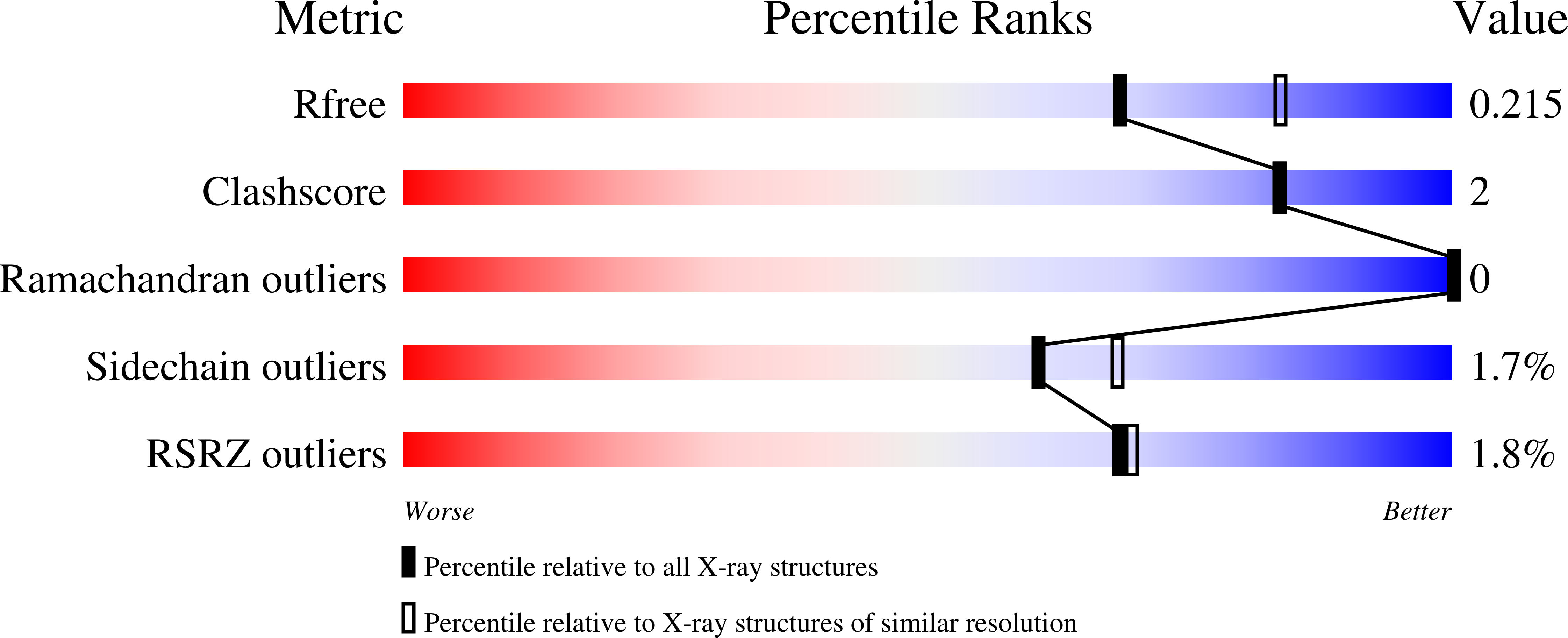
3QH2 - PubMed Abstract:
In many bacteria tenI is found clustered with genes involved in thiamin thiazole biosynthesis. However, while TenI shows high sequence similarity with thiamin phosphate synthase, the purified protein has no thiamin phosphate synthase activity, and the role of this enzyme in thiamin biosynthesis remains unknown. In this contribution, we identify the function of TenI as a thiazole tautomerase, describe the structure of the enzyme complexed with its reaction product, identify the substrates phosphate and histidine 122 as the acid/base residues involved in catalysis, and propose a mechanism for the reaction. The identification of the function of TenI completes the identification of all of the enzymes needed for thiamin biosynthesis by the major bacterial pathway.
Organizational Affiliation:
Department of Chemistry, Texas A&M University, College Station, Texas 77843, USA.