Combined SAXS/EM Based Models of the S. elongatus Post-Translational Circadian Oscillator and its Interactions with the Output His-Kinase SasA.
Pattanayek, R., Williams, D.R., Rossi, G., Weigand, S., Mori, T., Johnson, C.H., Stewart, P.L., Egli, M.(2011) PLoS One 6: e23697-e23697
- PubMed: 21887298
- DOI: https://doi.org/10.1371/journal.pone.0023697
- Primary Citation of Related Structures:
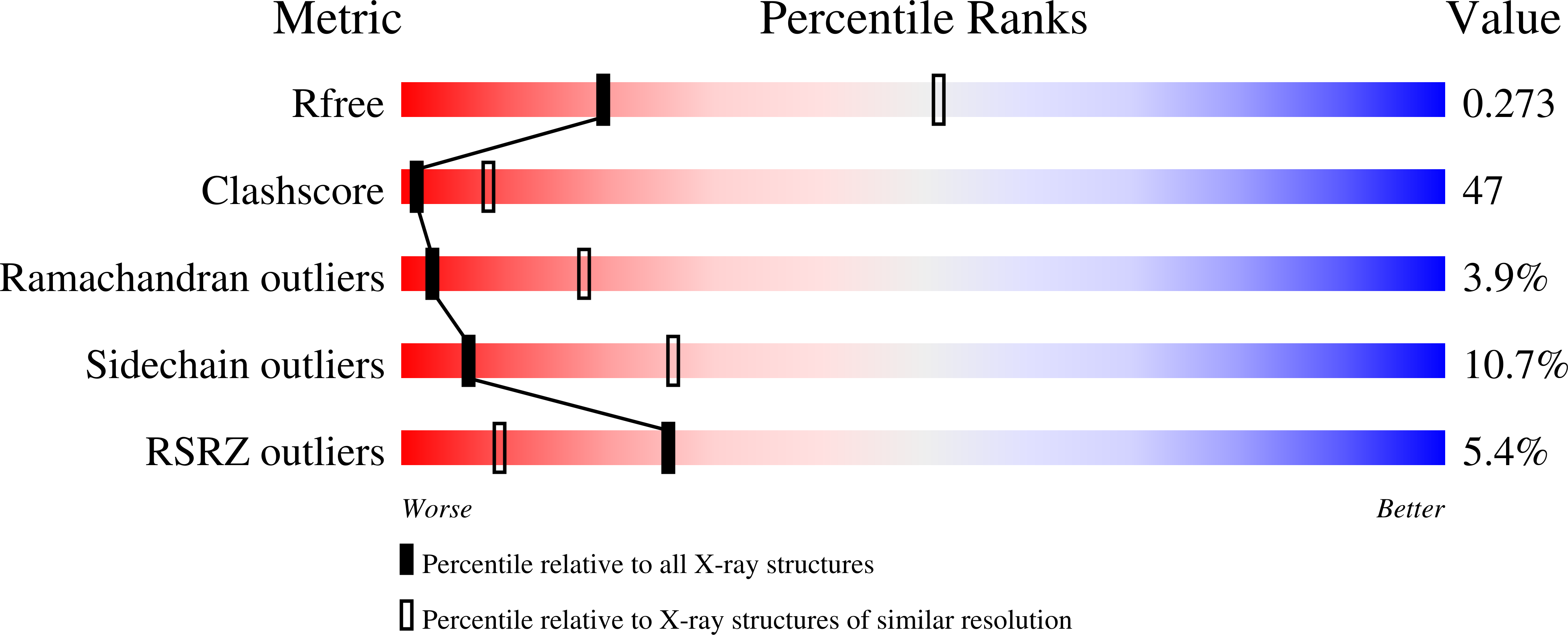
3S1A - PubMed Abstract:
The circadian clock in the cyanobacterium Synechococcus elongatus is composed of a post-translational oscillator (PTO) that can be reconstituted in vitro from three different proteins in the presence of ATP and a transcription-translation feedback loop (TTFL). The homo-hexameric KaiC kinase, phosphatase and ATPase alternates between hypo- and hyper-phosphorylated states over the 24-h cycle, with KaiA enhancing phosphorylation, and KaiB antagonizing KaiA and promoting KaiC subunit exchange. SasA is a His kinase that relays output signals from the PTO formed by the three Kai proteins to the TTFL. Although the crystal structures for all three Kai proteins are known, atomic resolution structures of Kai and Kai/SasA protein complexes have remained elusive. Here, we present models of the KaiAC and KaiBC complexes derived from solution small angle X-ray scattering (SAXS), which are consistent with previous EM based models. We also present a combined SAXS/EM model of the KaiC/SasA complex, which has two N-terminal SasA sensory domains occupying positions on the C-terminal KaiC ring reminiscent of the orientations adopted by KaiB dimers. Using EM we demonstrate that KaiB and SasA compete for similar binding sites on KaiC. We also propose an EM based model of the ternary KaiABC complex that is consistent with the sequestering of KaiA by KaiB on KaiC during the PTO dephosphorylation phase. This work provides the first 3D-catalogue of protein-protein interactions in the KaiABC PTO and the output pathway mediated by SasA.
Organizational Affiliation:
Department of Biochemistry, School of Medicine, Vanderbilt University, Nashville, Tennessee, United States of America.