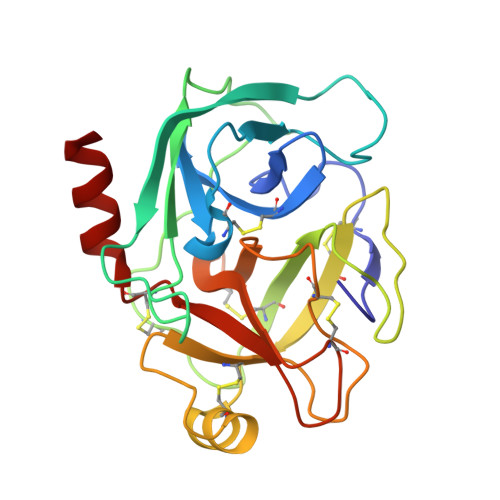
Harnessing the evolvability of tricyclic microviridins to dissect protease-inhibitor interactions.
Weiz, A.R., Ishida, K., Quitterer, F., Meyer, S., Kehr, J.C., Muller, K.M., Groll, M., Hertweck, C., Dittmann, E.(2014) Angew Chem Int Ed Engl 53: 3735-3738
- PubMed: 24591244
- DOI: https://doi.org/10.1002/anie.201309721
- Primary Citation of Related Structures:
4KTS, 4KTU - PubMed Abstract:
Understanding and controlling proteolysis is an important goal in therapeutic chemistry. Among the natural products specifically inhibiting proteases microviridins are particularly noteworthy. Microviridins are ribosomally produced and posttranslationally modified peptides that are processed into a unique, cagelike architecture. Here, we report a combined rational and random mutagenesis approach that provides fundamental insights into selectivity-conferring moieties of microviridins. The potent variant microviridin J was co-crystallized with trypsin, and for the first time the three-dimensional structure of microviridins was determined and the mode of inhibition revealed.
Organizational Affiliation:
Institute of Biochemistry and Biology, University of Potsdam, Karl-Liebknecht-Strasse 24-25, 14476 Potsdam-Golm (Germany).