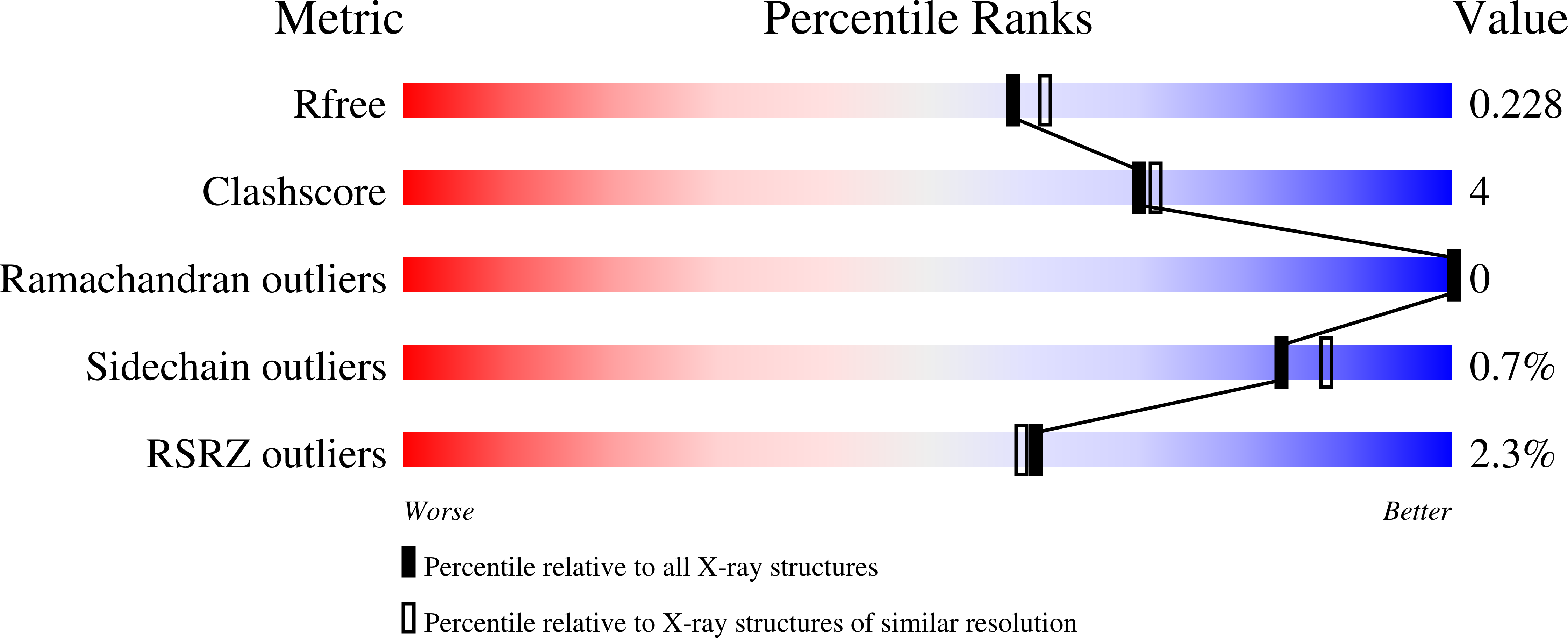
Structure of human farnesyl pyrophosphate synthase in complex with an aminopyridine bisphosphonate and two molecules of inorganic phosphate.
Park, J., Lin, Y.S., Tsantrizos, Y.S., Berghuis, A.M.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 299-304
- PubMed: 24598914
- DOI: https://doi.org/10.1107/S2053230X14002106
- Primary Citation of Related Structures:
4LFV - PubMed Abstract:
Human farnesyl pyrophosphate synthase (hFPPS) produces farnesyl pyrophosphate, an isoprenoid essential for a variety of cellular processes. The enzyme has been well established as the molecular target of the nitrogen-containing bisphosphonates (N-BPs), which are best known for their antiresorptive effects in bone but are also known for their anticancer properties. Crystal structures of hFPPS in ternary complexes with a novel bisphosphonate, YS0470, and the secondary ligands inorganic phosphate (Pi), inorganic pyrophosphate (PPi) and isopentenyl pyrophosphate (IPP) have recently been reported. Only the co-binding of the bisphosphonate with either PPi or IPP resulted in the full closure of the C-terminal tail of the enzyme, a conformational change that is required for catalysis and that is also responsible for the potent in vivo efficacy of N-BPs. In the present communication, a co-crystal structure of hFPPS in complex with YS0470 and two molecules of Pi is reported. The unusually close proximity between these ligands, which was confirmed by anomalous diffraction data, suggests that they interact with one another, with their anionic charges neutralized in their bound state. The structure also showed the tail of the enzyme to be fully disordered, indicating that simultaneous binding of two Pi molecules with a bisphosphonate cannot induce the tail-closing conformational change in hFPPS. Examination of homologous FPPSs suggested that this ligand-dependent tail closure is only conserved in the mammalian proteins. The prevalence of Pi-bound hFPPS structures in the PDB raises a question regarding the in vivo relevance of Pi binding to the function of the enzyme.
Organizational Affiliation:
Department of Biochemistry, McGill University, 3655 Promenade Sir William Osler, Montreal, QC H3G 1Y6, Canada.