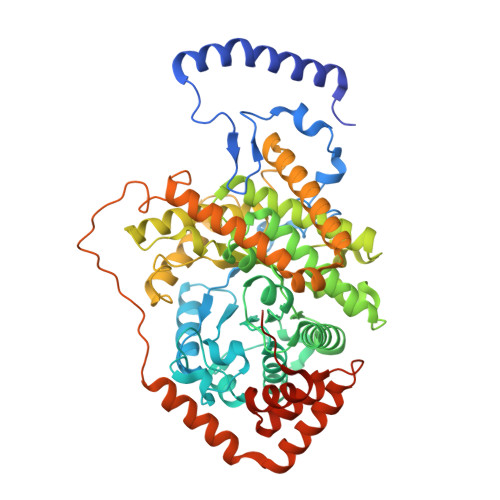
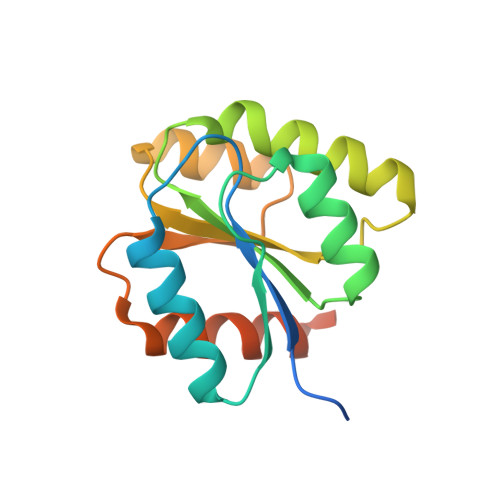
Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase.
Kurteva-Yaneva, N., Zahn, M., Weichler, M.T., Starke, R., Harms, H., Muller, R.H., Strater, N., Rohwerder, T.(2015) J Biol Chem 290: 9727-9737
- PubMed: 25720495
- DOI: https://doi.org/10.1074/jbc.M115.645689
- Primary Citation of Related Structures:
4R3U - PubMed Abstract:
Bacterial coenzyme B12-dependent 2-hydroxyisobutyryl-CoA mutase (HCM) is a radical enzyme catalyzing the stereospecific interconversion of (S)-3-hydroxybutyryl- and 2-hydroxyisobutyryl-CoA. It consists of two subunits, HcmA and HcmB. To characterize the determinants of substrate specificity, we have analyzed the crystal structure of HCM from Aquincola tertiaricarbonis in complex with coenzyme B12 and the substrates (S)-3-hydroxybutyryl- and 2-hydroxyisobutyryl-CoA in alternative binding. When compared with the well studied structure of bacterial and mitochondrial B12-dependent methylmalonyl-CoA mutase (MCM), HCM has a highly conserved domain architecture. However, inspection of the substrate binding site identified amino acid residues not present in MCM, namely HcmA Ile(A90) and Asp(A117). Asp(A117) determines the orientation of the hydroxyl group of the acyl-CoA esters by H-bond formation, thus determining stereospecificity of catalysis. Accordingly, HcmA D117A and D117V mutations resulted in significantly increased activity toward (R)-3-hydroxybutyryl-CoA. Besides interconversion of hydroxylated acyl-CoA esters, wild-type HCM as well as HcmA I90V and I90A mutant enzymes could also isomerize pivalyl- and isovaleryl-CoA, albeit at >10 times lower rates than the favorite substrate (S)-3-hydroxybutyryl-CoA. The nonconservative mutation HcmA D117V, however, resulted in an enzyme showing high activity toward pivalyl-CoA. Structural requirements for binding and isomerization of highly branched acyl-CoA substrates such as 2-hydroxyisobutyryl- and pivalyl-CoA, possessing tertiary and quaternary carbon atoms, respectively, are discussed.
Organizational Affiliation:
From the Department of Environmental Microbiology, Helmholtz Centre for Environmental Research (UFZ), 04318 Leipzig and.