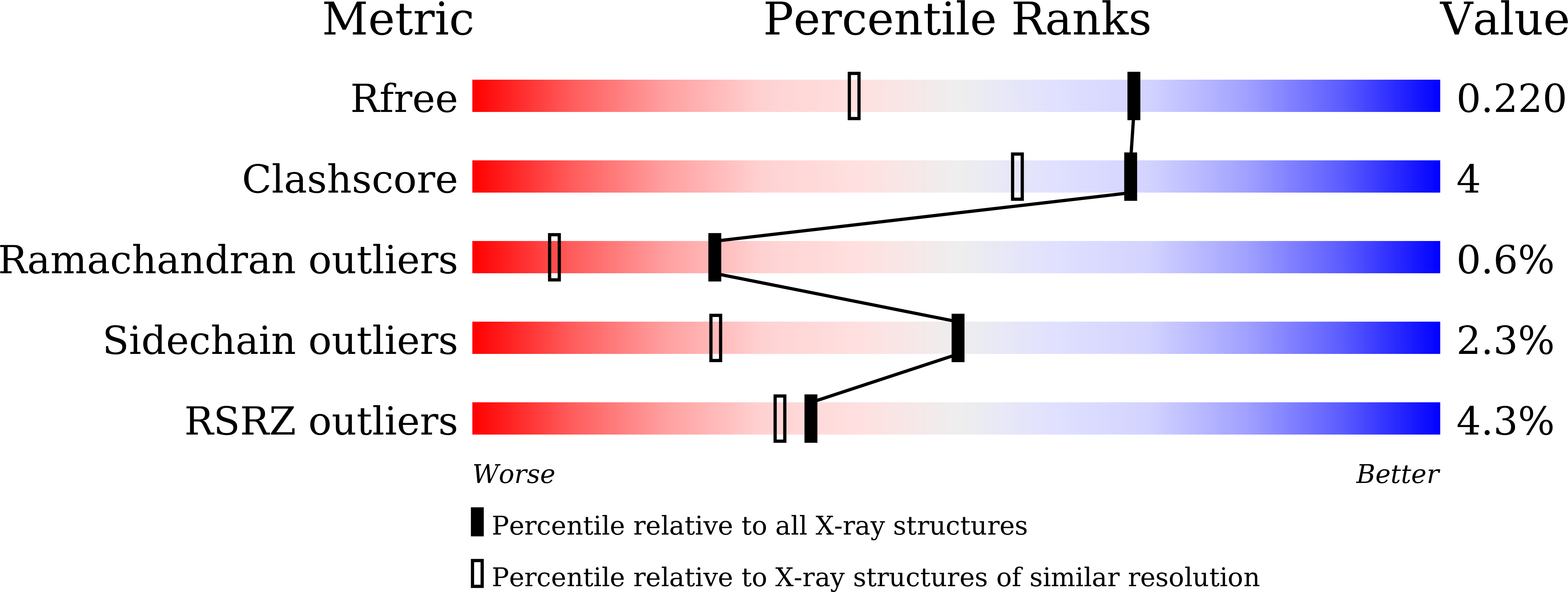
Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Hattie, M., Ito, T., Debowski, A.W., Arakawa, T., Katayama, T., Yamamoto, K., Fushinobu, S., Stubbs, K.A.(2015) Chem Commun (Camb) 51: 15008-15011
- PubMed: 26312778
- DOI: https://doi.org/10.1039/c5cc05494j
- Primary Citation of Related Structures:
5BXP, 5BXR, 5BXS, 5BXT - PubMed Abstract:
The synthesis of potent inhibitors for lacto-N-biosidases and X-ray structural characterization of these compounds in complex with BbLNBase is described.
Organizational Affiliation:
School of Chemistry and Biochemistry, The University of Western Australia, Crawley, WA 6009, Australia. [email protected].