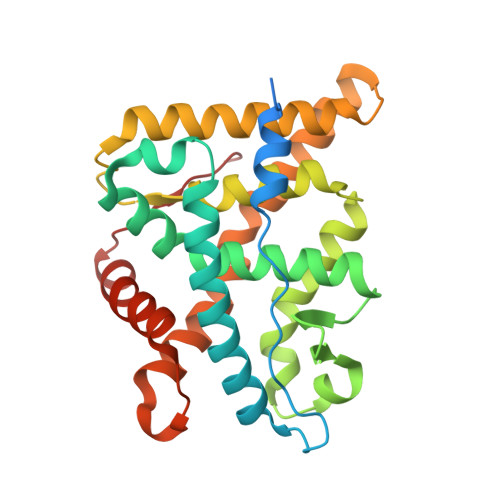
Discovery of New Selective Glucocorticoid Receptor Agonist Leads.
Berger, M., Rehwinkel, H., Schmees, N., Schacke, H., Edman, K., Wissler, L., Reichel, A., Jaroch, S.(2017) Bioorg Med Chem Lett 27: 437
- PubMed: 28043796
- DOI: https://doi.org/10.1016/j.bmcl.2016.12.047
- Primary Citation of Related Structures:
5G3J - PubMed Abstract:
We report on the discovery of two new lead series for the development of glucocorticoid receptor agonists. Firstly, the discovery of tetrahydronaphthalenes led to metabolically stable and dissociated compounds. Their binding mode to the glucocorticoid receptor could be elucidated through an X-ray structure. Closer inspection into the reaction path and analyses of side products revealed a new amino alcohol series also addressing the glucocorticoid receptor and demonstrating strong anti-inflammatory activity in vitro.
Organizational Affiliation:
Medicinal Chemistry Berlin, Candidate Generation & External Innovation, Drug Discovery, Bayer Pharma AG, D-13353 Berlin, Germany. Electronic address: [email protected].