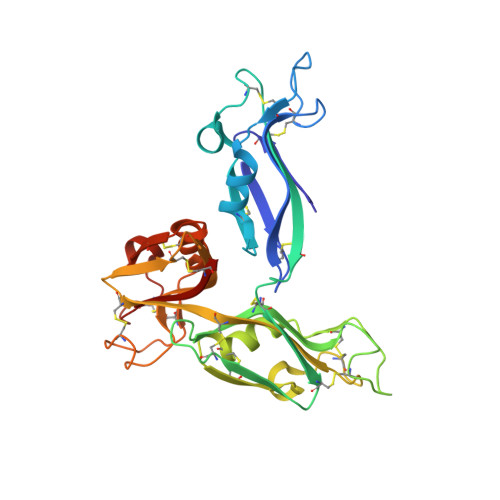
Carbohydrate recognition by the rhamnose-binding lectin SUL-I with a novel three-domain structure isolated from the venom of globiferous pedicellariae of the flower sea urchin Toxopneustes pileolus
Hatakeyama, T., Ichise, A., Unno, H., Goda, S., Oda, T., Tateno, H., Hirabayashi, J., Sakai, H., Nakagawa, H.(2017) Protein Sci 26: 1574-1583
- PubMed: 28470711
- DOI: https://doi.org/10.1002/pro.3185
- Primary Citation of Related Structures:
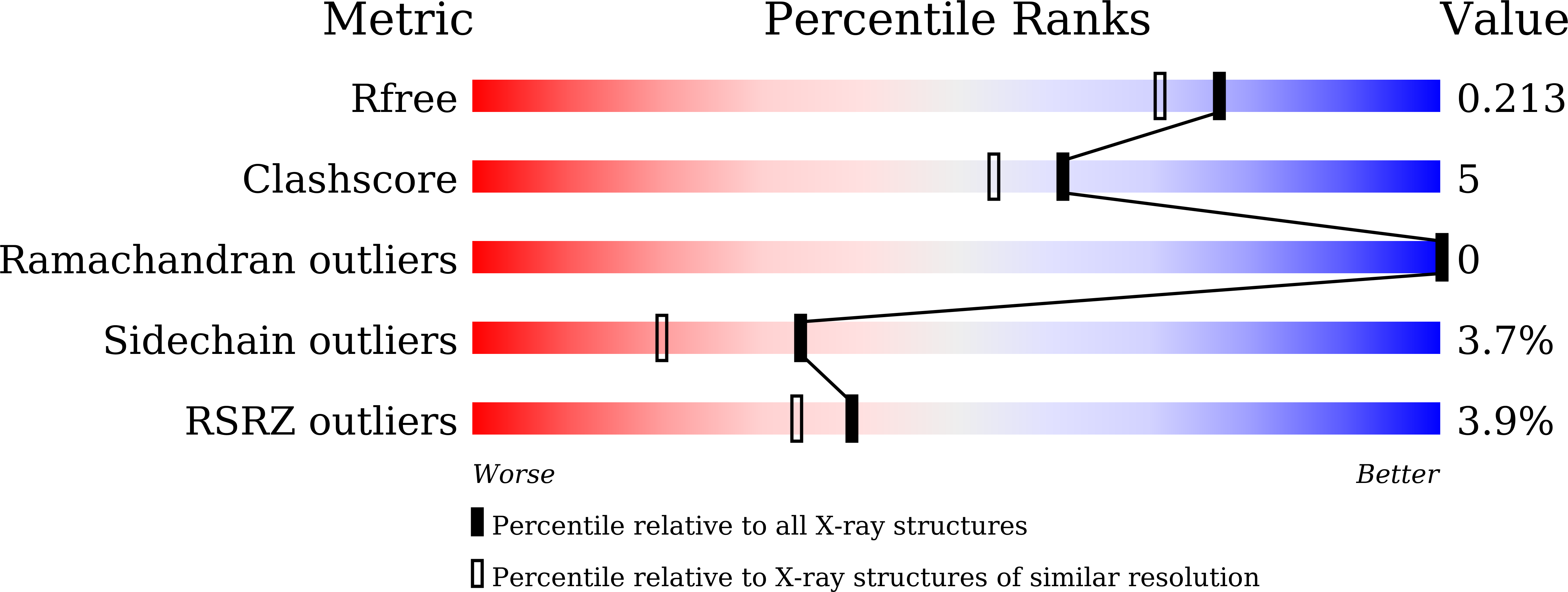
5H4S - PubMed Abstract:
The globiferous pedicellariae of the venomous sea urchin Toxopneustes pileolus contains several biologically active proteins. We have cloned the cDNA of one of the toxin components, SUL-I, which is a rhamnose-binding lectin (RBL) that acts as a mitogen through binding to carbohydrate chains on target cells. Recombinant SUL-I (rSUL-I) was produced in Escherichia coli cells, and its carbohydrate-binding specificity was examined with the glycoconjugate microarray analysis, which suggested that potential target carbohydrate structures are galactose-terminated N-glycans. rSUL-I exhibited mitogenic activity for murine splenocyte cells and toxicity against Vero cells. The three-dimensional structure of the rSUL-I/l-rhamnose complex was determined by X-ray crystallographic analysis at a 1.8 Å resolution. The overall structure of rSUL-I is composed of three distinctive domains with a folding structure similar to those of CSL3, a RBL from chum salmon (Oncorhynchus keta) eggs. The bound l-rhamnose molecules are mainly recognized by rSUL-I through hydrogen bonds between its 2-, 3-, and 4-hydroxy groups and Asp, Asn, and Glu residues in the binding sites, while Tyr and Ser residues participate in the recognition mechanism. It was also inferred that SUL-I may form a dimer in solution based on the molecular size estimated via dynamic light scattering as well as possible contact regions in its crystal structure.
Organizational Affiliation:
Biomolecular Chemistry Laboratory, Graduate School of Engineering, Nagasaki University, 852-8521, Japan.