Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4 beta-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin.
Zhao, W., Zhou, C., Guan, Z.Y., Yin, P., Chen, F., Tang, Y.J.(2017) ACS Chem Biol 12: 746-752
- PubMed: 28035796
- DOI: https://doi.org/10.1021/acschembio.6b00842
- Primary Citation of Related Structures:
5JCB - PubMed Abstract:
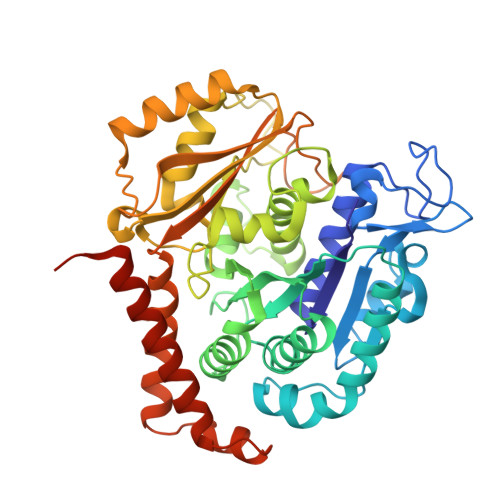
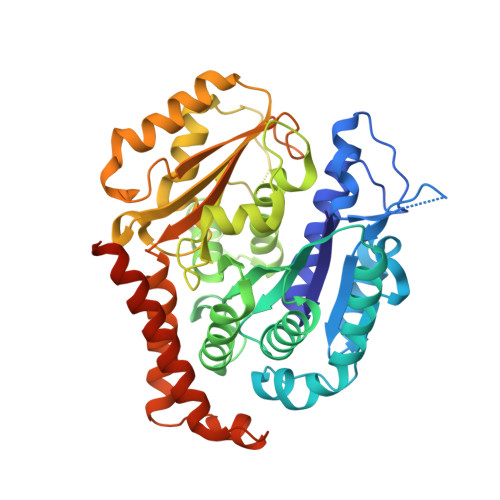
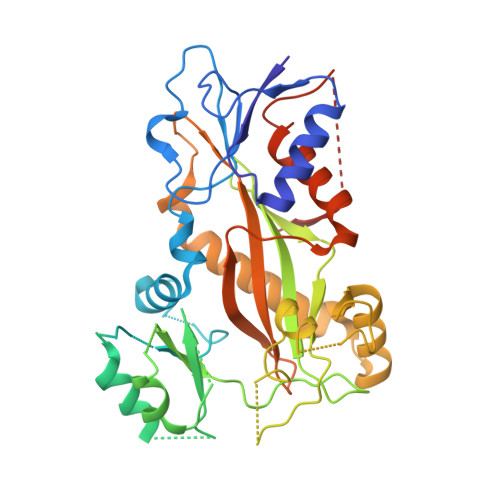
The colchicine domain is widely recognized as the binding site of microtubule depolymerization agents for anticancer drug design. Almost all of the drugs targeting the colchicine domain have been confirmed to bind to the tubulin β-subunit. Here we studied a crystal structure (2.3 Å) of the complex between tubulin and 4β-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin (compound 1S) with superior antitumor activity, which was designed on the basis of the colchicine domain and synthesized in our previous work. A distinct binding model of the colchicine domain was found in the complexes of tubulin with compound 1S. From a comparison of the crystal structures of tubulin-compound 1S and tubulin-colchicine complexes, the side chains of the T7 loop of β-tubulin flip outward and the T5 loop of α-tubulin changes its conformation. It has been shown that the β-subunit T7 loop reversibly participates in resistance to straightening that opposes microtubule assembly by flipping in and out. Together with the biochemical results from compound 1S, the structural data highlight the main contributors in the α-subunits and the colchicine domain β-subunits: the dual-target binding sites in the α-T7 loop and β-H7-T7 loop of tubulin. Compound 1S can synchronously bind to αβ-tubulin. The structures also highlight common features for the design and development of novel potent microtubule destabilizing agents.
Organizational Affiliation:
National Key Laboratory of Agromicrobiology, College of Food Sciences and Technology, Huazhong Agricultural University , Wuhan 430070, China.