Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
Taban, I.M., Elshihawy, H.E.A.E., Torun, B., Zucchini, B., Williamson, C.J., Altuwairigi, D., Ngu, A.S.T., McLean, K.J., Levy, C.W., Sood, S., Marino, L.B., Munro, A.W., de Carvalho, L.P.S., Simons, C.(2017) J Med Chem 60: 10257-10267
- PubMed: 29185746
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01562
- Primary Citation of Related Structures:
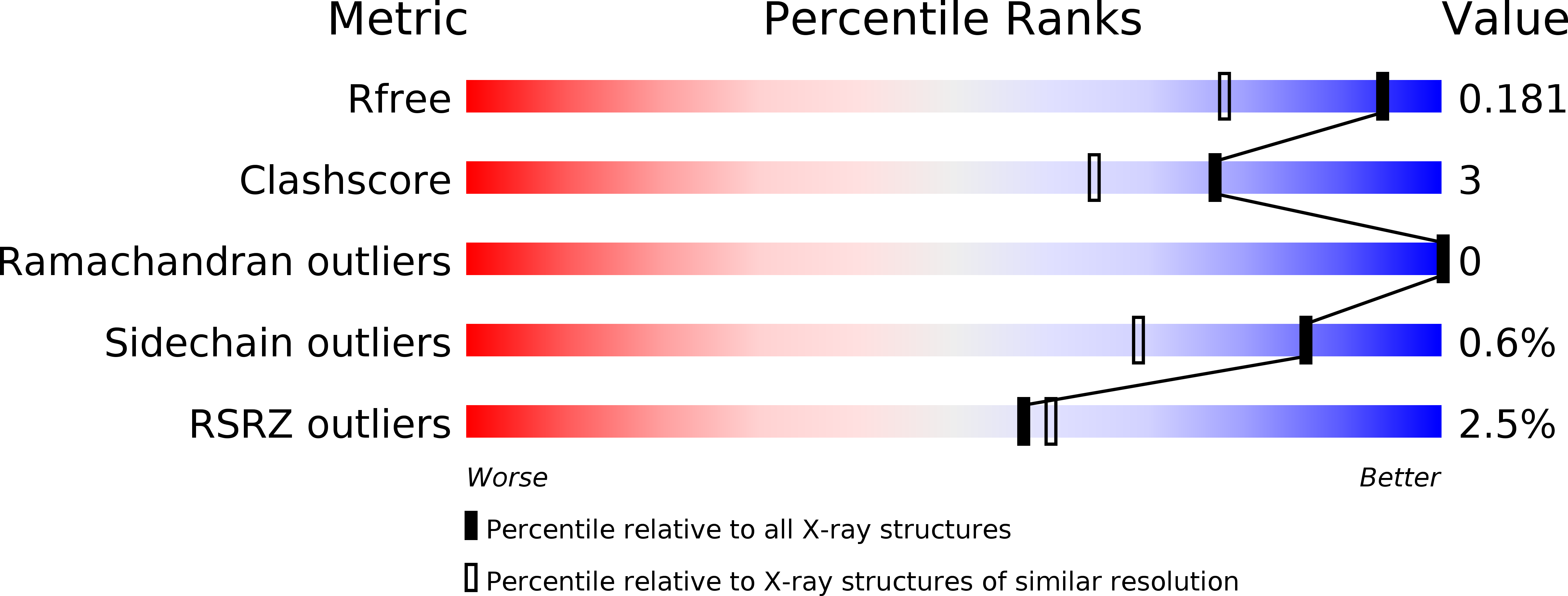
5O4K, 5O4L, 5OP9, 5OPA - PubMed Abstract:
Three series of biarylpyrazole imidazole and triazoles are described, which vary in the linker between the biaryl pyrazole and imidazole/triazole group. The imidazole and triazole series with the short -CH 2 - linker displayed promising antimycobacterial activity, with the imidazole-CH 2 - series (7) showing low MIC values (6.25-25 μg/mL), which was also influenced by lipophilicity. Extending the linker to -C(O)NH(CH 2 ) 2 - resulted in a loss of antimycobacterial activity. The binding affinity of the compounds with CYP121A1 was determined by UV-visible optical titrations with K D values of 2.63, 35.6, and 290 μM, respectively, for the tightest binding compounds 7e, 8b, and 13d from their respective series. Both binding affinity assays and docking studies of the CYP121A1 inhibitors suggest type II indirect binding through interstitial water molecules, with key binding residues Thr77, Val78, Val82, Val83, Met86, Ser237, Gln385, and Arg386, comparable with the binding interactions observed with fluconazole and the natural substrate dicyclotyrosine.
Organizational Affiliation:
School of Pharmacy & Pharmaceutical Sciences, Cardiff University , King Edward VII Avenue, Cardiff CF10 3NB, U.K.