Conformational Switch Regulates the DNA Cytosine Deaminase Activity of Human APOBEC3B.
Shi, K., Demir, O., Carpenter, M.A., Wagner, J., Kurahashi, K., Harris, R.S., Amaro, R.E., Aihara, H.(2017) Sci Rep 7: 17415-17415
- PubMed: 29234087
- DOI: https://doi.org/10.1038/s41598-017-17694-3
- Primary Citation of Related Structures:
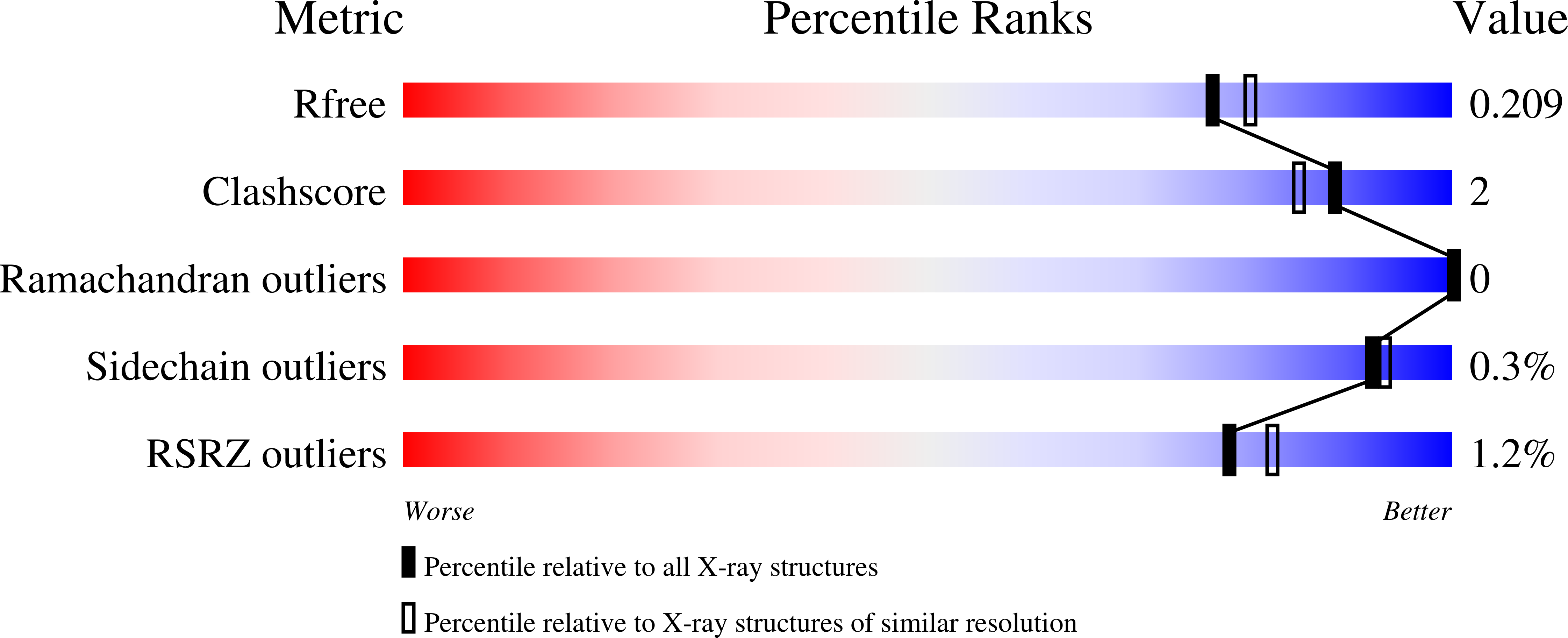
5SXG, 5SXH - PubMed Abstract:
The APOBEC3B (A3B) single-stranded DNA (ssDNA) cytosine deaminase has important roles in innate immunity but is also a major endogenous source of mutations in cancer. Previous structural studies showed that the C-terminal catalytic domain of human A3B has a tightly closed active site, and rearrangement of the surrounding loops is required for binding to substrate ssDNA. Here we report structures of the A3B catalytic domain in a new crystal form that show alternative, yet still closed, conformations of active site loops. All-atom molecular dynamics simulations support the dynamic behavior of active site loops and recapitulate the distinct modes of interactions that maintain a closed active site. Replacing segments of A3B loop 1 to mimic the more potent cytoplasmic deaminase APOBEC3A leads to elevated ssDNA deaminase activity, likely by facilitating opening of the active site. These data collectively suggest that conformational equilibrium of the A3B active site loops, skewed toward being closed, controls enzymatic activity by regulating binding to ssDNA substrates.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, Minnesota, 55455, USA.