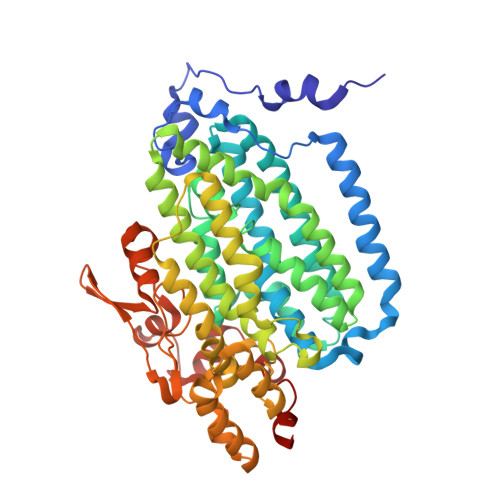
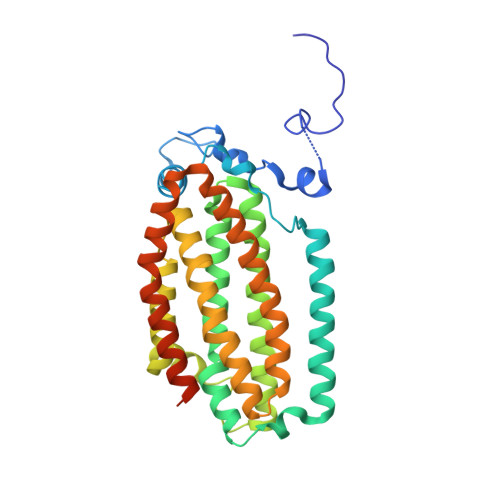
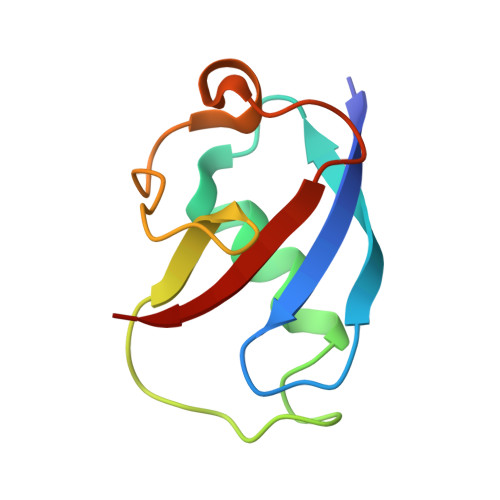
In-crystal reaction cycle of a toluene-bound diiron hydroxylase.
Acheson, J.F., Bailey, L.J., Brunold, T.C., Fox, B.G.(2017) Nature 544: 191-195
- PubMed: 28346937
- DOI: https://doi.org/10.1038/nature21681
- Primary Citation of Related Structures:
5TDS, 5TDT, 5TDU, 5TDV - PubMed Abstract:
Electrophilic aromatic substitution is one of the most important and recognizable classes of organic chemical transformation. Enzymes create the strong electrophiles that are needed for these highly energetic reactions by using O 2 , electrons, and metals or other cofactors. Although the nature of the oxidants that carry out electrophilic aromatic substitution has been deduced from many approaches, it has been difficult to determine their structures. Here we show the structure of a diiron hydroxylase intermediate formed during a reaction with toluene. Density functional theory geometry optimizations of an active site model reveal that the intermediate is an arylperoxo Fe 2+ /Fe 3+ species with delocalized aryl radical character. The structure suggests that a carboxylate ligand of the diiron centre may trigger homolytic cleavage of the O-O bond by transferring a proton from a metal-bound water. Our work provides the spatial and electronic constraints needed to propose a comprehensive mechanism for diiron enzyme arene hydroxylation that accounts for many prior experimental results.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA.