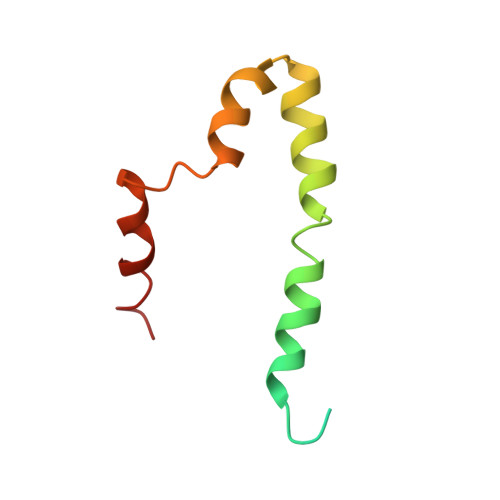
Structural model of the SARS coronavirus E channel in LMPG micelles
Surya, W., Li, Y., Torres, J.(2018) Biochim Biophys Acta 1860: 1309-1317
- PubMed: 29474890
- DOI: https://doi.org/10.1016/j.bbamem.2018.02.017
- Primary Citation of Related Structures:
5X29 - PubMed Abstract:
Coronaviruses (CoV) cause common colds in humans, but are also responsible for the recent Severe Acute, and Middle East, respiratory syndromes (SARS and MERS, respectively). A promising approach for prevention are live attenuated vaccines (LAVs), some of which target the envelope (E) protein, which is a small membrane protein that forms ion channels. Unfortunately, detailed structural information is still limited for SARS-CoV E, and non-existent for other CoV E proteins. Herein, we report a structural model of a SARS-CoV E construct in LMPG micelles with, for the first time, unequivocal intermolecular NOEs. The model corresponding to the detergent-embedded region is consistent with previously obtained orientational restraints obtained in lipid bilayers and in vivo escape mutants. The C-terminal domain is mostly α-helical, and extramembrane intermolecular NOEs suggest interactions that may affect the TM channel conformation.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551, Singapore.