Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marineVibriobacteria.
Suginta, W., Sritho, N., Ranok, A., Bulmer, D.M., Kitaoku, Y., van den Berg, B., Fukamizo, T.(2018) J Biol Chem 293: 5150-5159
- PubMed: 29444825
- DOI: https://doi.org/10.1074/jbc.RA117.001012
- Primary Citation of Related Structures:
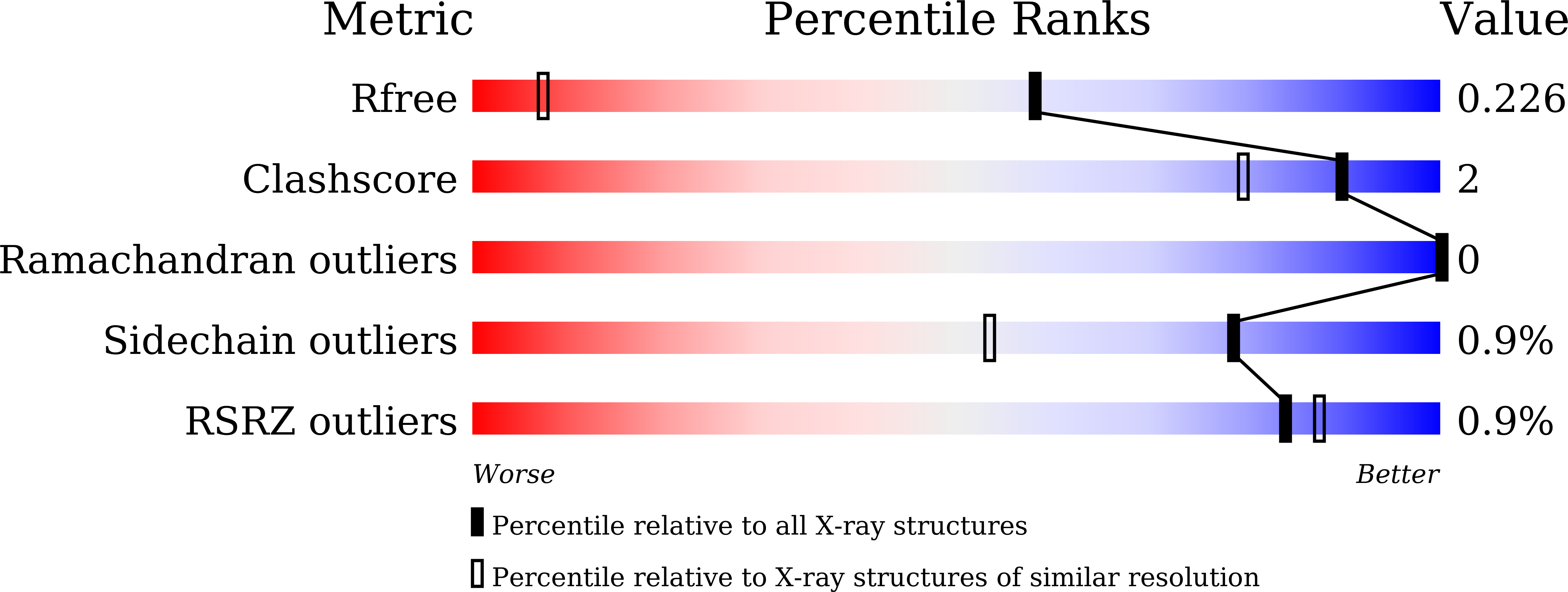
5YQW - PubMed Abstract:
Periplasmic solute-binding proteins in bacteria are involved in the active transport of nutrients into the cytoplasm. In marine bacteria of the genus Vibrio , a chitooligosaccharide-binding protein (CBP) is thought to be the major solute-binding protein controlling the rate of chitin uptake in these bacteria. However, the molecular mechanism of the CBP involvement in chitin metabolism has not been elucidated. Here, we report the structure and function of a recombinant chitooligosaccharide-binding protein from Vibrio harveyi , namely Vh CBP, expressed in Escherichia coli Isothermal titration calorimetry revealed that Vh CBP strongly binds shorter chitooligosaccharides ((GlcNAc) n , where n = 2, 3, and 4) with affinities that are considerably greater than those for glycoside hydrolase family 18 and 19 chitinases but does not bind longer ones, including insoluble chitin polysaccharides. We also found that VhCBP comprises two domains with flexible linkers and that the domain-domain interface forms the sugar-binding cleft, which is not long extended but forms a small cavity. (GlcNAc) 2 bound to this cavity, apparently triggering a closed conformation of Vh CBP. Trp-363 and Trp-513, which stack against the two individual GlcNAc rings, likely make a major contribution to the high affinity of Vh CBP for (GlcNAc) 2 The strong chitobiose binding, followed by the conformational change of Vh CBP, may facilitate its interaction with an active-transport system in the inner membrane of Vibrio species.
Organizational Affiliation:
From the Biochemistry-Electrochemistry Research Unit, Institute of Science, Suranaree University of Technology, Nakhon Ratchasima 30000, Thailand, [email protected].