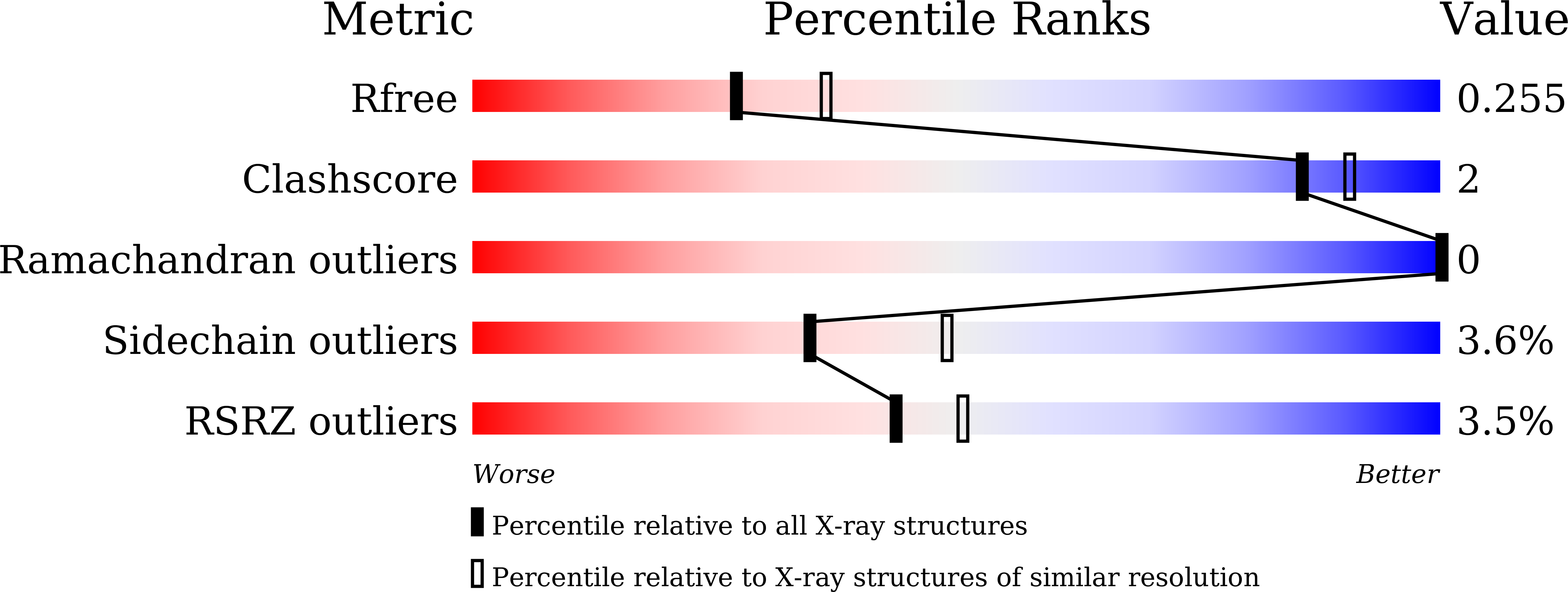
Identifying purine nucleoside phosphorylase as the target of quinine using cellular thermal shift assay.
Dziekan, J.M., Yu, H., Chen, D., Dai, L., Wirjanata, G., Larsson, A., Prabhu, N., Sobota, R.M., Bozdech, Z., Nordlund, P.(2019) Sci Transl Med 11
- PubMed: 30602534
- DOI: https://doi.org/10.1126/scitranslmed.aau3174
- Primary Citation of Related Structures:
5ZNC, 5ZNI - PubMed Abstract:
Mechanisms of action (MoAs) have been elusive for most antimalarial drugs in clinical use. Decreasing responsiveness to antimalarial treatments stresses the need for a better resolved understanding of their MoAs and associated resistance mechanisms. In the present work, we implemented the cellular thermal shift assay coupled with mass spectrometry (MS-CETSA) for drug target identification in Plasmodium falciparum , the main causative agent of human malaria. We validated the efficacy of this approach for pyrimethamine, a folic acid antagonist, and E64d, a broad-spectrum cysteine proteinase inhibitor. Subsequently, we applied MS-CETSA to quinine and mefloquine, two important antimalarial drugs with poorly characterized MoAs. Combining studies in the P. falciparum parasite lysate and intact infected red blood cells, we found P. falciparum purine nucleoside phosphorylase (PfPNP) as a common binding target for these two quinoline drugs. Biophysical and structural studies with a recombinant protein further established that both compounds bind within the enzyme's active site. Quinine binds to PfPNP at low nanomolar affinity, suggesting a substantial contribution to its therapeutic effect. Overall, we demonstrated that implementation of MS-CETSA for P. falciparum constitutes a promising strategy to elucidate the MoAs of existing and candidate antimalarial drugs.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 637551, Singapore.