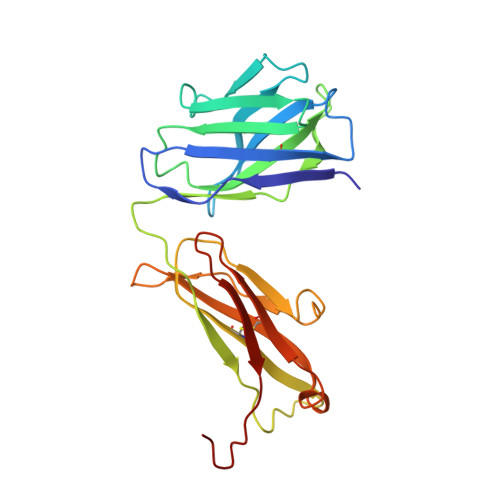
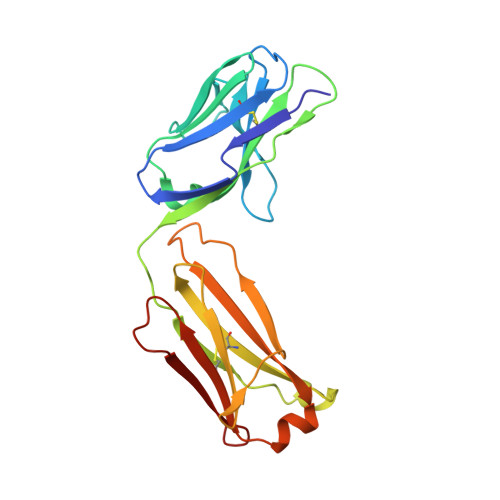
Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch.
Schinagl, A., Kerschbaumer, R.J., Sabarth, N., Douillard, P., Scholz, P., Voelkel, D., Hollerweger, J.C., Goettig, P., Brandstetter, H., Scheiflinger, F., Thiele, M.(2018) Biochemistry 57: 1523-1532
- PubMed: 29412660
- DOI: https://doi.org/10.1021/acs.biochem.7b01156
- Primary Citation of Related Structures:
6FOE - PubMed Abstract:
Macrophage migration inhibitory factor (MIF) is a pro-inflammatory and tumor-promoting cytokine that occurs in two redox-dependent immunologically distinct conformational isoforms. The disease-related structural isoform of MIF (oxMIF) can be specifically and predominantly detected in the circulation of patients with inflammatory diseases and in tumor tissue, whereas the ubiquitously expressed isoform of MIF (redMIF) is abundantly expressed in healthy and diseased subjects. In this article, we report that cysteine 81 within MIF serves as a "switch cysteine" for the conversion of redMIF to oxMIF. Modulating cysteine 81 by thiol reactive agents leads to significant structural rearrangements of the protein, resulting in a decreased β-sheet content and an increased random coil content, but maintaining the trimeric quaternary structure. This conformational change in the MIF molecule enables binding of oxMIF-specific antibodies BaxB01 and BaxM159, which showed beneficial activity in animal models of inflammation and cancer. Crystal structure analysis of the MIF-derived EPCALCS peptide, bound in its oxMIF-like conformation by the Fab fragment of BaxB01, revealed that this peptide adopts a curved conformation, making the central thiol protein oxidoreductase motif competent to undergo disulfide shuffling. We conclude that redMIF might reflect a latent zymogenic form of MIF, and formation of oxMIF leads to a physiologically relevant, i.e., enzymatically active, state.
Organizational Affiliation:
Baxalta Innovations GmbH , Uferstrasse 15 , 2304 Orth an der Donau , Austria.