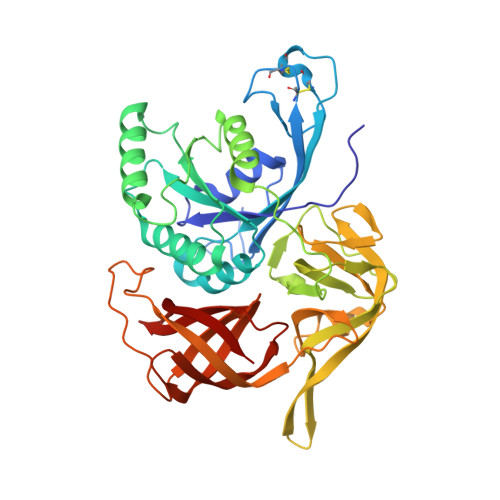
The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Nikonov, O., Kravchenko, O., Nevskaya, N., Stolboushkina, E., Garber, M., Nikonov, S.(2019) Acta Crystallogr D Struct Biol 75: 392-399
- PubMed: 30988256
- DOI: https://doi.org/10.1107/S2059798319002304
- Primary Citation of Related Structures:
6H6K, 6I5M - PubMed Abstract:
The structure of the γ subunit of archaeal translation initiation factor 2 (aIF2) from Sulfolobus solfataricus (SsoIF2γ) was determined in complex with GDPCP (a GTP analog). Crystals were obtained in the absence of magnesium ions in the crystallization solution. They belonged to space group P1, with five molecules in the unit cell. Four of these molecules are related in pairs by a common noncrystallographic twofold symmetry axis, while the fifth has no symmetry equivalent. Analysis of the structure and its comparison with other known aIF2 γ-subunit structures in the GTP-bound state show that (i) the magnesium ion is necessary for the formation and the maintenance of the active form of SsoIF2γ and (ii) in addition to the two previously known structural switches 1 and 2, eukaryotic translation initiation factor 2 (eIF2) and aIF2 molecules have another flexible region (switch 3), the function of which may consist of initiation of the hydrolysis of GTP and the removal of e/aIF2 from the ribosome after codon-anticodon recognition.
Organizational Affiliation:
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Russian Federation.