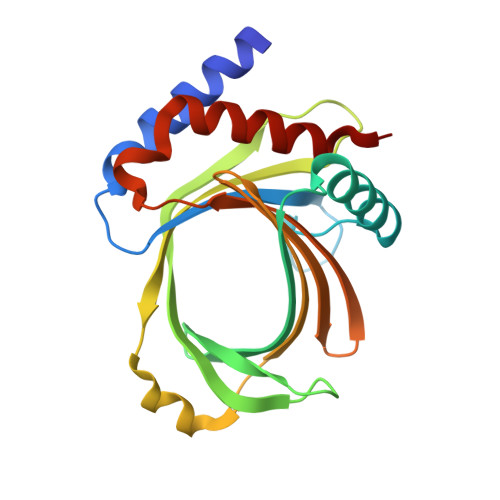
Crystal structures of the RNA triphosphatase fromTrypanosoma cruziprovide insights into how it recognizes the 5'-end of the RNA substrate.
Takagi, Y., Kuwabara, N., Dang, T.T., Furukawa, K., Ho, C.K.(2020) J Biol Chem 295: 9076-9086
- PubMed: 32381506
- DOI: https://doi.org/10.1074/jbc.RA119.011811
- Primary Citation of Related Structures:
6L7V, 6L7W, 6L7X, 6L7Y - PubMed Abstract:
RNA triphosphatase catalyzes the first step in mRNA cap formation, hydrolysis of the terminal phosphate from the nascent mRNA transcript. The RNA triphosphatase from the protozoan parasite Trypanosoma cruzi , TcCet1, belongs to the family of triphosphate tunnel metalloenzymes (TTMs). TcCet1 is a promising antiprotozoal drug target because the mechanism and structure of the protozoan RNA triphosphatases are completely different from those of the RNA triphosphatases found in mammalian and arthropod hosts. Here, we report several crystal structures of the catalytically active form of TcCet1 complexed with a divalent cation and an inorganic tripolyphosphate in the active-site tunnel at 2.20-2.51 Å resolutions. The structures revealed that the overall structure, the architecture of the tunnel, and the arrangement of the metal-binding site in TcCet1 are similar to those in other TTM proteins. On the basis of the position of three sulfate ions that cocrystallized on the positively charged surface of the protein and results obtained from mutational analysis, we identified an RNA-binding site in TcCet1. We conclude that the 5'-end of the triphosphate RNA substrate enters the active-site tunnel directionally. The structural information reported here provides valuable insight into designing inhibitors that could specifically block the entry of the triphosphate RNA substrate into the TTM-type RNA triphosphatases of T. cruzi and related pathogens.
Organizational Affiliation:
Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, Tsukuba, Ibaraki, Japan.