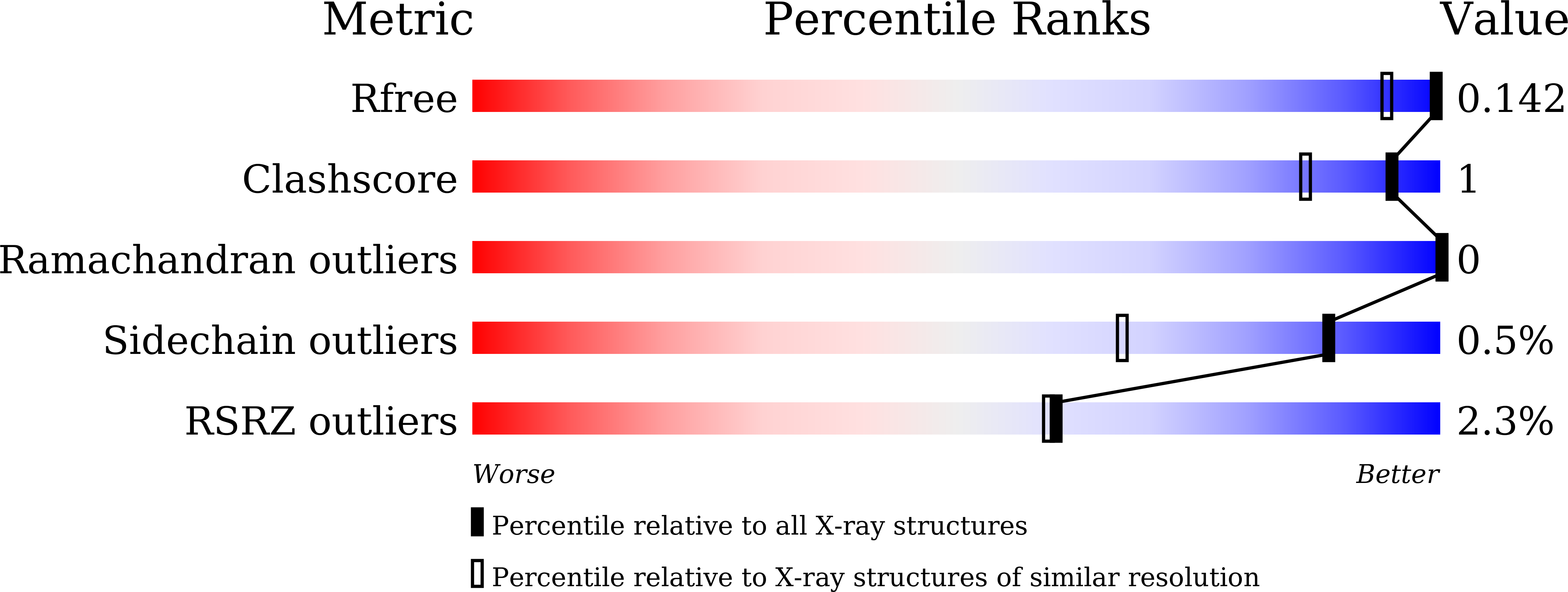
Elucidating the role of metal ions in carbonic anhydrase catalysis.
Kim, J.K., Lee, C., Lim, S.W., Adhikari, A., Andring, J.T., McKenna, R., Ghim, C.M., Kim, C.U.(2020) Nat Commun 11: 4557-4557
- PubMed: 32917908
- DOI: https://doi.org/10.1038/s41467-020-18425-5
- Primary Citation of Related Structures:
6LUU, 6LUV, 6LUW, 6LUX, 6LUY, 6LUZ, 6LV1, 6LV2, 6LV3, 6LV4, 6LV5, 6LV6, 6LV7, 6LV8, 6LV9, 6LVA - PubMed Abstract:
Why metalloenzymes often show dramatic changes in their catalytic activity when subjected to chemically similar but non-native metal substitutions is a long-standing puzzle. Here, we report on the catalytic roles of metal ions in a model metalloenzyme system, human carbonic anhydrase II (CA II). Through a comparative study on the intermediate states of the zinc-bound native CA II and non-native metal-substituted CA IIs, we demonstrate that the characteristic metal ion coordination geometries (tetrahedral for Zn 2+ , tetrahedral to octahedral conversion for Co 2+ , octahedral for Ni 2+ , and trigonal bipyramidal for Cu 2+ ) directly modulate the catalytic efficacy. In addition, we reveal that the metal ions have a long-range (~10 Å) electrostatic effect on restructuring water network in the active site. Our study provides evidence that the metal ions in metalloenzymes have a crucial impact on the catalytic mechanism beyond their primary chemical properties.
Organizational Affiliation:
Department of Physics, Ulsan National Institute of Science and Technology (UNIST), Ulsan, 44919, Republic of Korea.