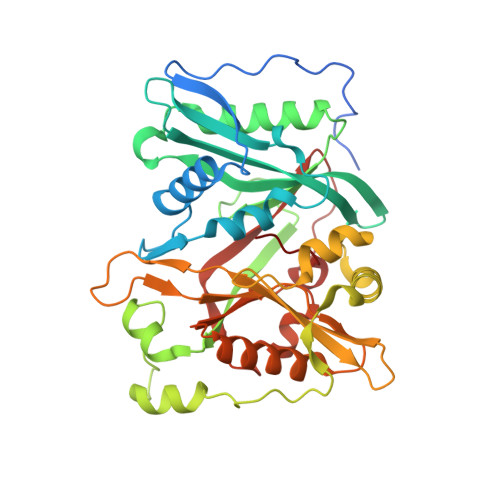
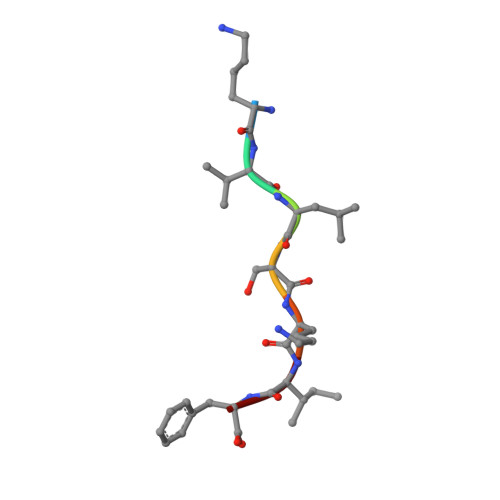
NMT1 and NMT2 are lysine myristoyltransferases regulating the ARF6 GTPase cycle.
Kosciuk, T., Price, I.R., Zhang, X., Zhu, C., Johnson, K.N., Zhang, S., Halaby, S.L., Komaniecki, G.P., Yang, M., DeHart, C.J., Thomas, P.M., Kelleher, N.L., Christopher Fromme, J., Lin, H.(2020) Nat Commun 11: 1067-1067
- PubMed: 32103017
- DOI: https://doi.org/10.1038/s41467-020-14893-x
- Primary Citation of Related Structures:
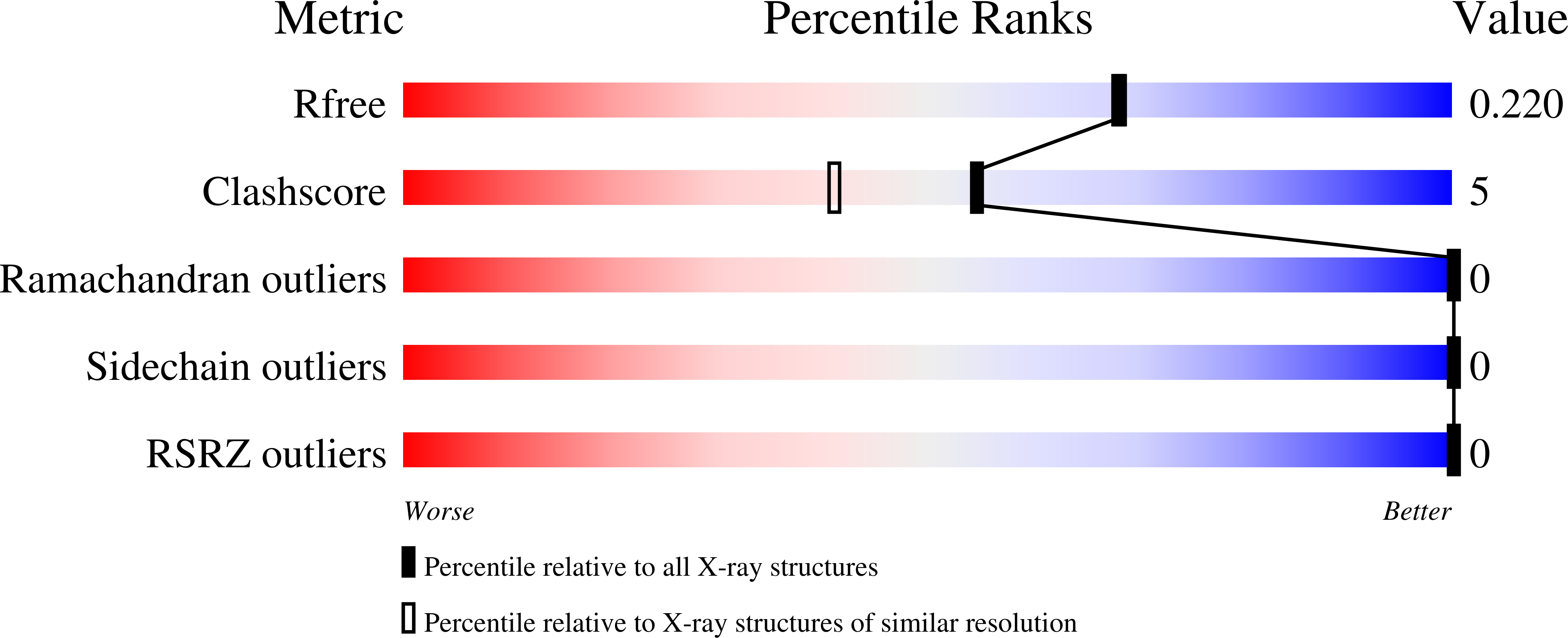
6PAU, 6PAV - PubMed Abstract:
Lysine fatty acylation in mammalian cells was discovered nearly three decades ago, yet the enzymes catalyzing it remain unknown. Unexpectedly, we find that human N-terminal glycine myristoyltransferases (NMT) 1 and 2 can efficiently myristoylate specific lysine residues. They modify ADP-ribosylation factor 6 (ARF6) on lysine 3 allowing it to remain on membranes during the GTPase cycle. We demonstrate that the NAD + -dependent deacylase SIRT2 removes the myristoyl group, and our evidence suggests that NMT prefers the GTP-bound while SIRT2 prefers the GDP-bound ARF6. This allows the lysine myrisotylation-demyristoylation cycle to couple to and promote the GTPase cycle of ARF6. Our study provides an explanation for the puzzling dissimilarity of ARF6 to other ARFs and suggests the existence of other substrates regulated by this previously unknown function of NMT. Furthermore, we identified a NMT/SIRT2-ARF6 regulatory axis, which may offer new ways to treat human diseases.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY, 14853, USA.