Novel N-substituted 5-phosphate-d-arabinonamide derivatives as strong inhibitors of phosphoglucose isomerases: Synthesis, structure-activity relationship and crystallographic studies.
Ahmad, L., Plancqueel, S., Lazar, N., Korri-Youssoufi, H., Li de la Sierra-Gallay, I., van Tilbeurgh, H., Salmon, L.(2020) Bioorg Chem 102: 104048-104048
- PubMed: 32682158
- DOI: https://doi.org/10.1016/j.bioorg.2020.104048
- Primary Citation of Related Structures:
6XUH, 6XUI - PubMed Abstract:
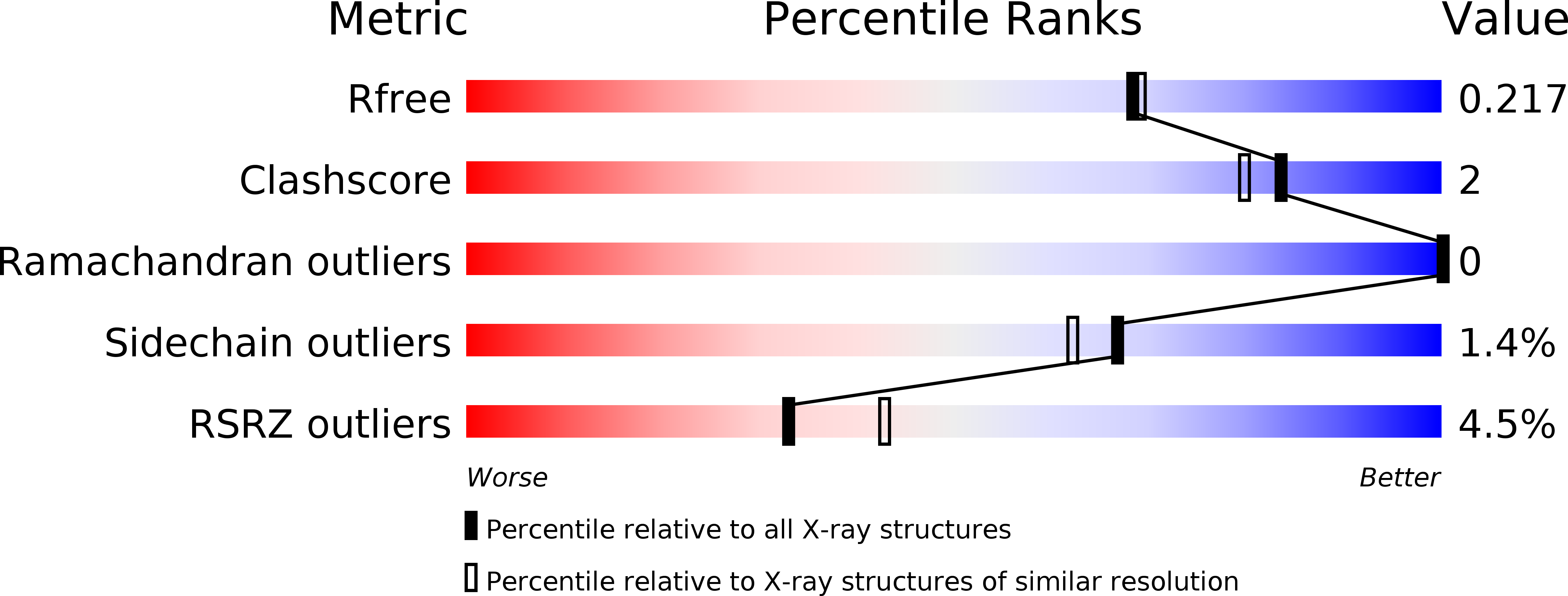
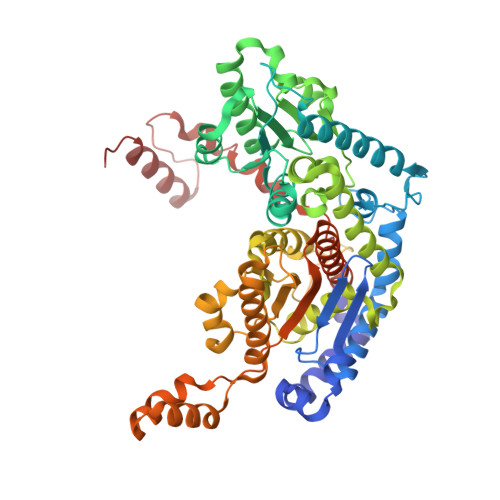
Phosphoglucose isomerase (PGI) is a cytosolic enzyme that catalyzes the reversible interconversion of d-glucose 6-phosphate and d-fructose 6-phosphate in glycolysis. Outside the cell, PGI is also known as autocrine motility factor (AMF), a cytokine secreted by a large variety of tumor cells that stimulates motility of cancer cells in vitro and metastases development in vivo. Human PGI and AMF are strictly identical proteins both in terms of sequence and 3D structure, and AMF activity is known to involve, at least in part, the enzymatic active site. Hence, with the purpose of finding new strong AMF-PGI inhibitors that could be potentially used as anticancer agents and/or as bioreceptors for carbohydrate-based electrochemical biosensors, we report in this study the synthesis and kinetic evaluation of several new human PGI inhibitors derived from the synthon 5-phospho-d-arabinono-1,4-lactone. Although not designed as high-energy intermediate analogue inhibitors of the enzyme catalyzed isomerization reaction, several of these N-substituted 5-phosphate-d-arabinonamide derivatives appears as new strong PGI inhibitors. For one of them, we report its crystal structure in complex with human PGI at 2.38 Å. Detailed analysis of its interactions at the active site reveals a new binding mode and shows that human PGI is relatively tolerant for modified inhibitors at the "head" C-1 part, offering promising perspectives for the future design of carbohydrate-based biosensors.
Organizational Affiliation:
Institut de Chimie Moléculaire et des Matériaux d'Orsay (ICMMO), Equipe de Chimie Bioorganique et Bioinorganique, CNRS UMR8182, LabEx LERMIT, Université Paris-Saclay, Rue du Doyen Georges Poitou, bât. 420, 91405 Orsay Cedex, France.