Fragment KCL_914 in complex with MAP kinase p38-alpha
De Nicola, G.F., Nichols, C.E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitogen-activated protein kinase 14 | 360 | Mus musculus | Mutation(s): 1 Gene Names: Mapk14, Crk1, Csbp1, Csbp2 EC: 2.7.11.24 | 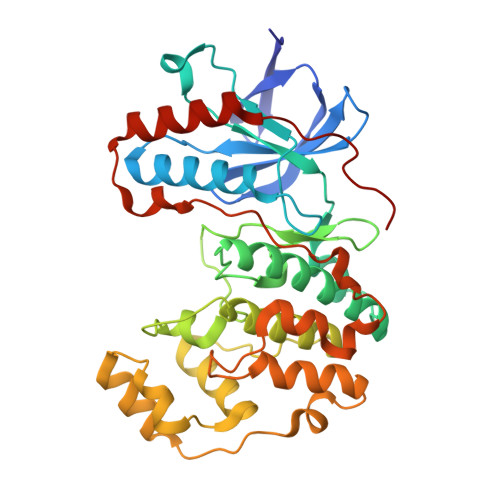 | |
UniProt | |||||
Find proteins for P47811 (Mus musculus) Explore P47811 Go to UniProtKB: P47811 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P47811 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SB4 Query on SB4 | C [auth A] | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE C18 H19 F N6 VSPFURGQAYMVAN-UHFFFAOYSA-N |  | ||
| OG2 (Subject of Investigation/LOI) Query on OG2 | B [auth A] | 1-(1-adamantyl)-3-ethyl-guanidine C13 H23 N3 VGBFQSVFIIHEDN-DNSLJTBWSA-N |  | ||
| SO4 Query on SO4 | I [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| CA Query on CA | D [auth A], E [auth A], F [auth A], G [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | H [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 80.732 | α = 90 |
| b = 102.7 | β = 90 |
| c = 103.733 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DIALS | data collection |
| PHENIX | refinement |
| DIALS | data reduction |
| DIALS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| British Heart Foundation | United Kingdom | SP/14/2/30922, FS/14/29/30896 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MC_PC_17164 |