Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Atkinson, E.L., Iegre, J., D'Amore, C., Brear, P., Salvi, M., Hyvonen, M., Spring, D.R.(2022) Chem Commun (Camb) 58: 4791-4794
- PubMed: 35343996
- DOI: https://doi.org/10.1039/d2cc00707j
- Primary Citation of Related Structures:
6YZH, 6Z19, 7QUX - PubMed Abstract:
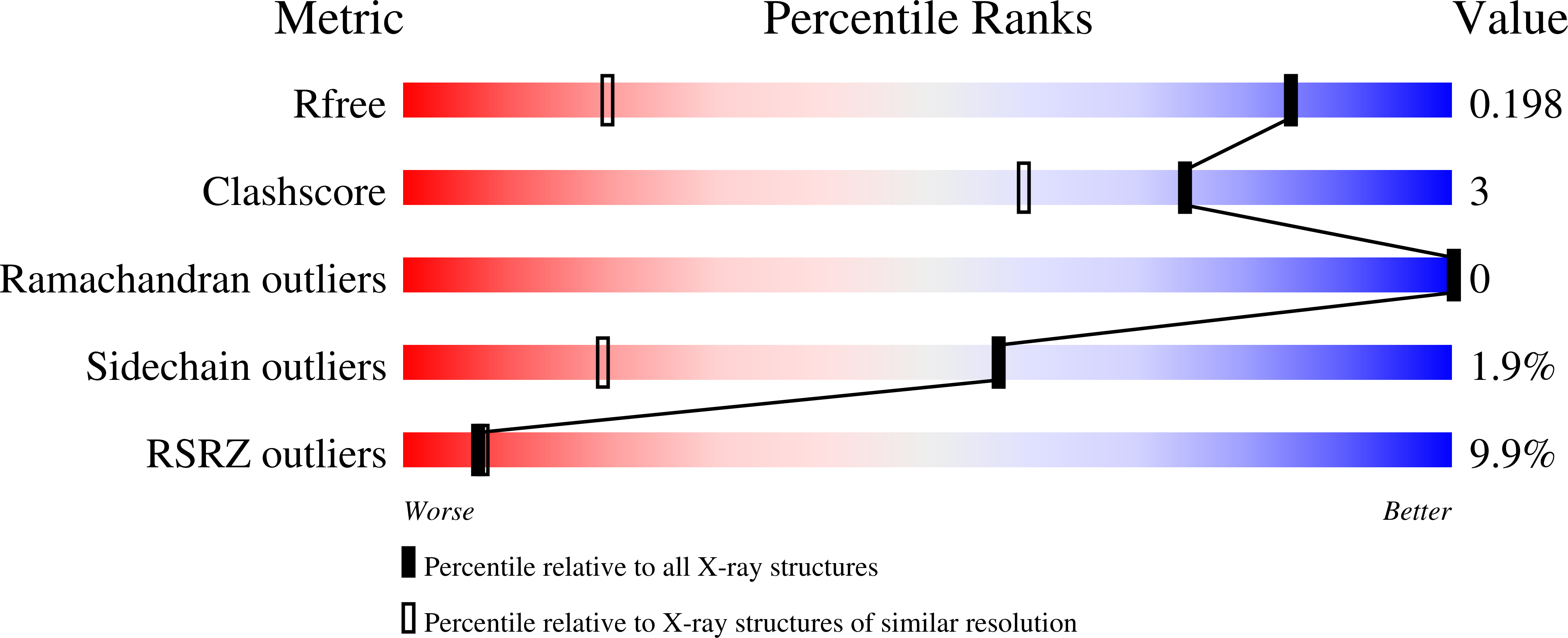
In this work, an iterative cycle of enzymatic assays, X-ray crystallography, molecular modelling and cellular assays were used to develop a functionalisable chemical probe for the CK2α/β PPI. The lead peptide, P8C9, successfully binds to CK2α at the PPI site, is easily synthesisable and functionalisable, highly stable in serum and small enough to accommodate further optimisation.
Organizational Affiliation:
Yusuf Hamied Department of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW, Cambridge, UK. [email protected].