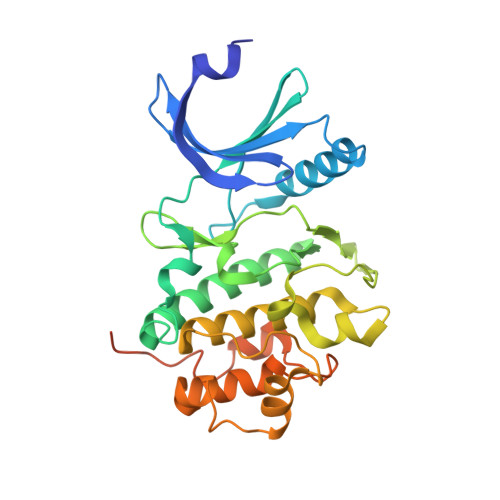
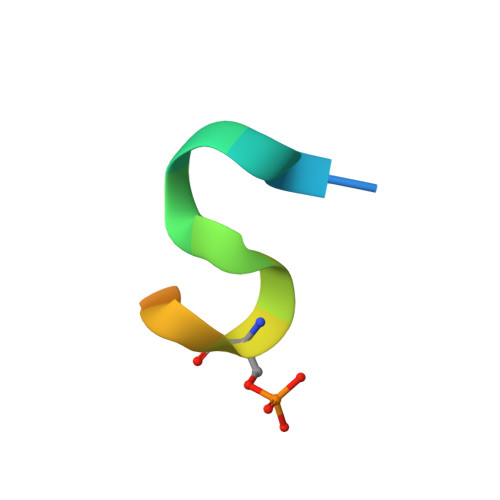Structural basis for recruitment of the CHK1 DNA damage kinase by the CLASPIN scaffold protein.
Day, M., Parry-Morris, S., Houghton-Gisby, J., Oliver, A.W., Pearl, L.H.(2021) Structure 29: 531
- PubMed: 33789090
- DOI: https://doi.org/10.1016/j.str.2021.03.007
- Primary Citation of Related Structures:
7AKM, 7AKO - PubMed Abstract:
CHK1 is a protein kinase that functions downstream of activated ATR to phosphorylate multiple targets as part of intra-S and G2/M DNA damage checkpoints. Its role in allowing cells to survive replicative stress has made it an important target for anti-cancer drug discovery. Activation of CHK1 by ATR depends on their mutual interaction with CLASPIN, a natively unstructured protein that interacts with CHK1 through a cluster of phosphorylation sites in its C-terminal half. We have now determined the crystal structure of the kinase domain of CHK1 bound to a high-affinity motif from CLASPIN. Our data show that CLASPIN engages a conserved site on CHK1 adjacent to the substrate-binding cleft, involved in phosphate sensing in other kinases. The CLASPIN motif is not phosphorylated by CHK1, nor does it affect phosphorylation of a CDC25 substrate peptide, suggesting that it functions purely as a scaffold for CHK1 activation by ATR.
Organizational Affiliation:
Cancer Research UK DNA Repair Enzymes Group, Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Falmer, Brighton BN1 9RQ, UK.