Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
Watanabe, M., Fujihara, M., Motoyama, T., Kawasaki, M., Yamada, S., Takamura, Y., Ito, S., Makishima, M., Nakano, S., Kakuta, H.(2021) J Med Chem 64: 430-439
- PubMed: 33356247
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01354
- Primary Citation of Related Structures:
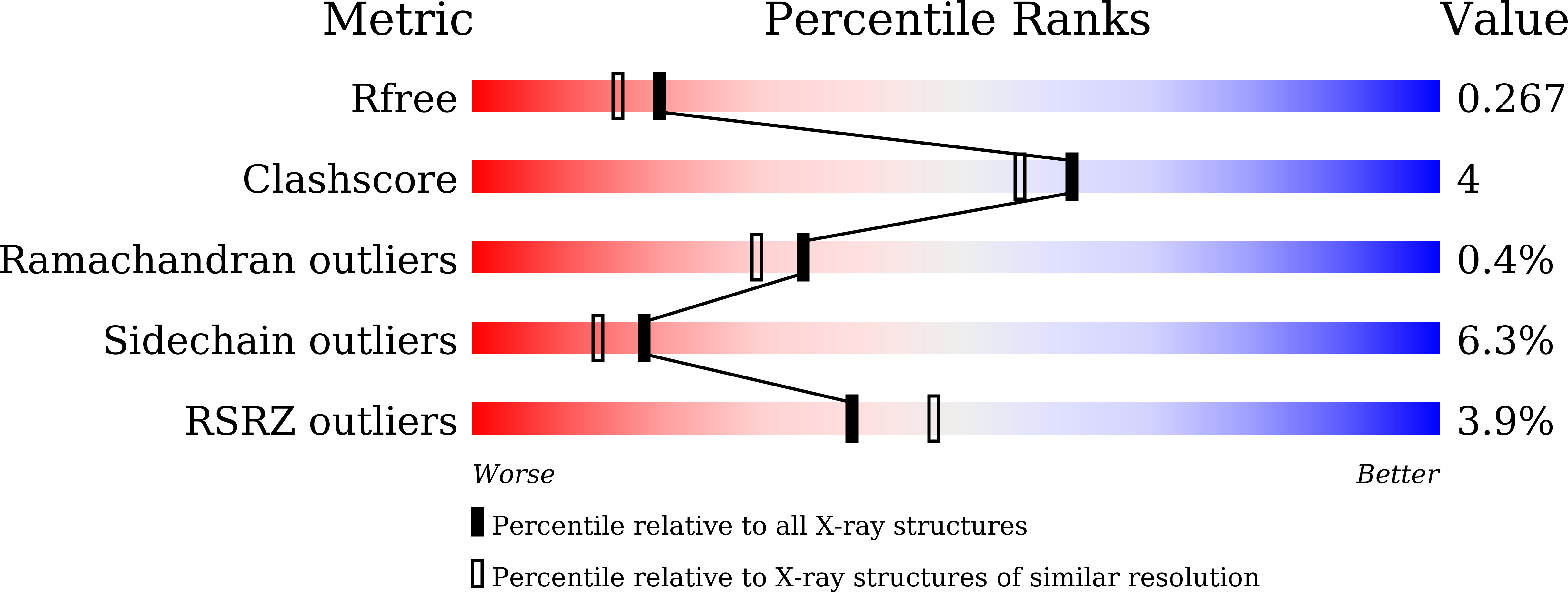
7CFO - PubMed Abstract:
Retinoid X receptor (RXR) heterodimers such as PPAR/RXR, LXR/RXR, and FXR/RXR can be activated by RXR agonists alone and are therefore designated as permissive. Similarly, existing RXR antagonists show allosteric antagonism toward partner receptor agonists in these permissive RXR heterodimers. Here, we show 1-(3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-2-(trifluoromethyl)-1 H -benzo[ d ]imidazole-5-carboxylic acid ( 14 , CBTF-EE) as the first RXR antagonist that does not show allosteric inhibition in permissive RXR heterodimers. This compound was designed based on the hypothesis that RXR antagonists that do not induce conformational changes of RXR would not exhibit such allosteric inhibition. CD spectra and X-ray co-crystallography of the complex of 14 and the RXR ligand binding domain (LBD) confirmed that 14 does not change the conformation of hRXR-LBD. The X-ray structure analysis revealed that 14 binds at the entrance of the ligand binding pocket (LBP), blocking access to the LBP and thus serving as a "gatekeeper".
Organizational Affiliation:
Division of Pharmaceutical Sciences, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, 1-1-1, Tsushima-naka, Kita-ku, Okayama 700-8530, Japan.