Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Watanabe-Takahashi, M., Tamada, M., Senda, M., Hibino, M., Shimizu, E., Okuta, A., Miyazawa, A., Senda, T., Nishikawa, K.(2021) Commun Biol 4: 538-538
- PubMed: 33972673
- DOI: https://doi.org/10.1038/s42003-021-02068-3
- Primary Citation of Related Structures:
7D6Q, 7D6R - PubMed Abstract:
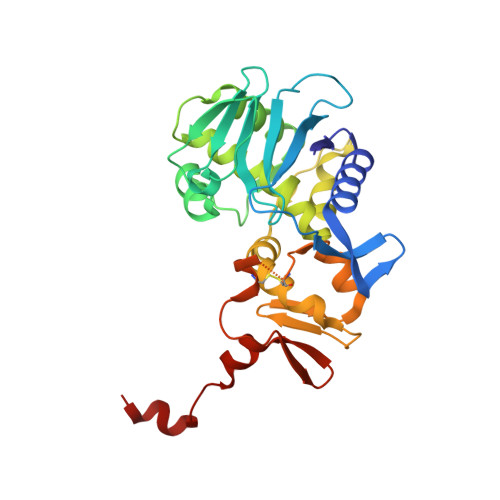
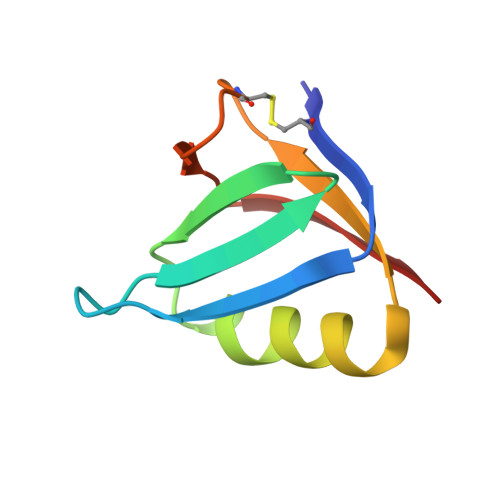
Shiga toxin (Stx) is a major virulence factor of enterohemorrhagic Escherichia coli, which causes fatal systemic complications. Here, we identified a tetravalent peptide that inhibited Stx by targeting its receptor-binding, B-subunit pentamer through a multivalent interaction. A monomeric peptide with the same motif, however, did not bind to the B-subunit pentamer. Instead, the monomer inhibited cytotoxicity with remarkable potency by binding to the catalytic A-subunit. An X-ray crystal structure analysis to 1.6 Å resolution revealed that the monomeric peptide fully occupied the catalytic cavity, interacting with Glu167 and Arg170, both of which are essential for catalytic activity. Thus, the peptide motif demonstrated potent inhibition of two functionally distinct subunits of Stx.
Organizational Affiliation:
Department of Molecular Life Sciences, Graduate School of Life and Medical Sciences, Doshisha University, Kyoto, Japan.