State-selective modulation of heterotrimeric G alpha s signaling with macrocyclic peptides.
Dai, S.A., Hu, Q., Gao, R., Blythe, E.E., Touhara, K.K., Peacock, H., Zhang, Z., von Zastrow, M., Suga, H., Shokat, K.M.(2022) Cell 185: 3950
- PubMed: 36170854
- DOI: https://doi.org/10.1016/j.cell.2022.09.019
- Primary Citation of Related Structures:
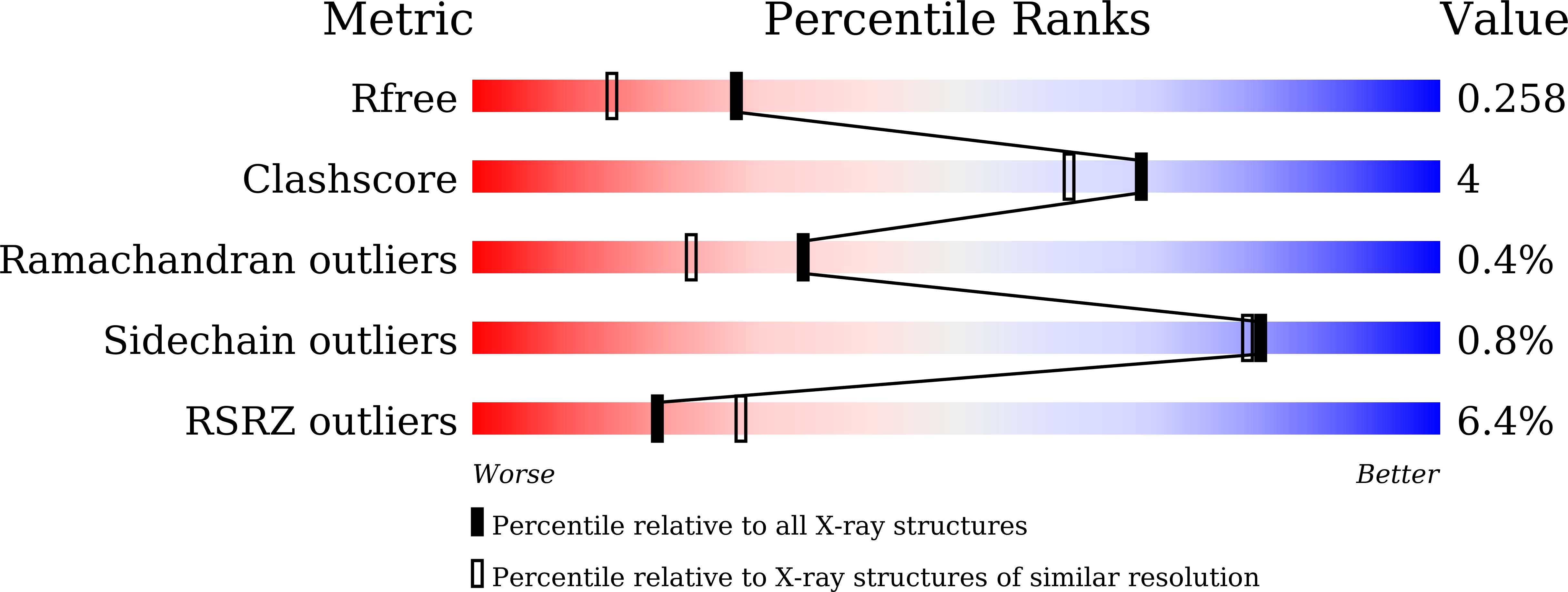
7BPH, 7E5E - PubMed Abstract:
The G protein-coupled receptor cascade leading to production of the second messenger cAMP is replete with pharmacologically targetable proteins, with the exception of the Gα subunit, Gαs. GTPases remain largely undruggable given the difficulty of displacing high-affinity guanine nucleotides and the lack of other drug binding sites. We explored a chemical library of 10 12 cyclic peptides to expand the chemical search for inhibitors of this enzyme class. We identified two macrocyclic peptides, GN13 and GD20, that antagonize the active and inactive states of Gαs, respectively. Both macrocyclic peptides fine-tune Gαs activity with high nucleotide-binding-state selectivity and G protein class-specificity. Co-crystal structures reveal that GN13 and GD20 distinguish the conformational differences within the switch II/α3 pocket. Cell-permeable analogs of GN13 and GD20 modulate Gαs/Gβγ signaling in cells through binding to crystallographically defined pockets. The discovery of cyclic peptide inhibitors targeting Gαs provides a path for further development of state-dependent GTPase inhibitors.
Organizational Affiliation:
Department of Cellular and Molecular Pharmacology, University of California, San Francisco, San Francisco, CA 94158, USA; Howard Hughes Medical Institute, Chevy Chase, MD 20815, USA.