Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy.
Blum, E., Zhang, J., Zaluski, J., Einstein, D.E., Korshin, E.E., Kubas, A., Gruzman, A., Tochtrop, G.P., Kiser, P.D., Palczewski, K.(2021) J Med Chem 64: 8287-8302
- PubMed: 34081480
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00279
- Primary Citation of Related Structures:
7K88, 7K89, 7K8G, 7L0E - PubMed Abstract:
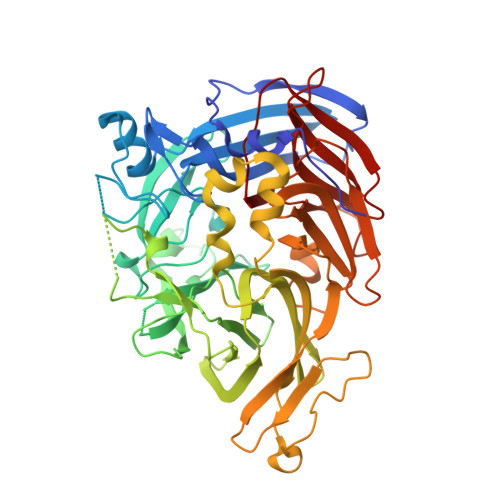
Recycling of all- trans -retinal to 11- cis -retinal through the visual cycle is a fundamental metabolic pathway in the eye. A potent retinoid isomerase (RPE65) inhibitor, ( R )-emixustat, has been developed and tested in several clinical trials; however, it has not received regulatory approval for use in any specific retinopathy. Rapid clearance of this drug presents challenges to maintaining concentrations in eyes within a therapeutic window. To address this pharmacokinetic inadequacy, we rationally designed and synthesized a series of emixustat derivatives with strategically placed fluorine and deuterium atoms to slow down the key metabolic transformations known for emixustat. Crystal structures and quantum chemical analysis of RPE65 in complex with the most potent emixustat derivatives revealed the structural and electronic bases for how fluoro substituents can be favorably accommodated within the active site pocket of RPE65. We found a close (∼3.0 Å) F-π interaction that is predicted to contribute ∼2.4 kcal/mol to the overall binding energy.
Organizational Affiliation:
Department of Chemistry, Faculty of Exact Sciences, Bar-Ilan University, Ramat-Gan 5290002, Israel.